Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN669-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN669-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 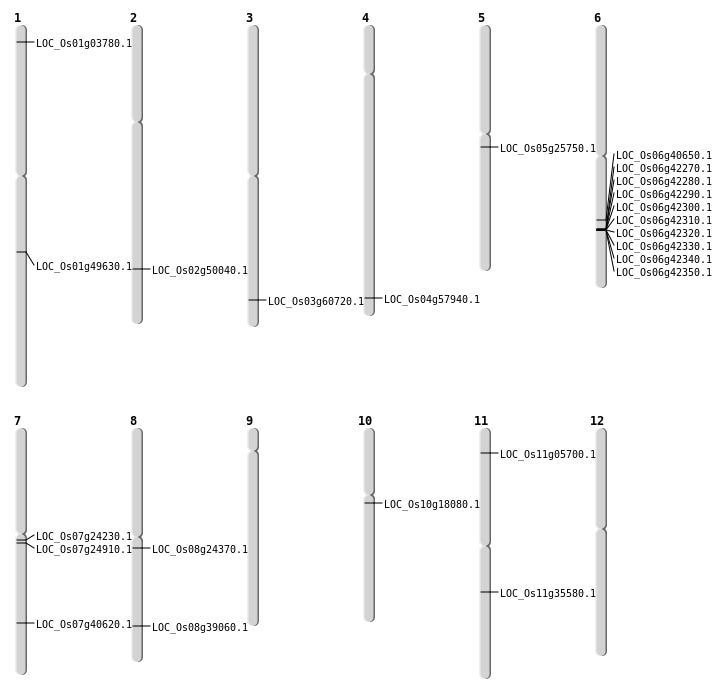
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11047376 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 1590162 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g03780 |
| SBS | Chr1 | 24565401 | T-C | HET | INTRON | |
| SBS | Chr1 | 41739187 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 12398847 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 2086008 | G-A | HET | INTRON | |
| SBS | Chr10 | 9131285 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 20862091 | C-A | HET | STOP_GAINED | LOC_Os11g35580 |
| SBS | Chr12 | 16156177 | A-T | HET | INTRON | |
| SBS | Chr12 | 16156178 | A-T | HET | INTRON | |
| SBS | Chr12 | 26063872 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 12112065 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 27730661 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 31219078 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 32988724 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 6228019 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 7228817 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 15211412 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 487107 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 34248420 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 3490779 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 10763663 | T-A | HET | INTRON | |
| SBS | Chr6 | 12487683 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 30436203 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 10255522 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 13778195 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g24230 |
| SBS | Chr7 | 1701214 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 20783800 | G-A | HET | INTRON | |
| SBS | Chr7 | 27266193 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 13777220 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 24688920 | T-A | HET | INTRON | |
| SBS | Chr8 | 2630308 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 3073656 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 18624055 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 4010583 | A-T | HET | SYNONYMOUS_CODING |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 891173 | 891175 | 2 | |
| Deletion | Chr11 | 2606967 | 2607010 | 43 | LOC_Os11g05700 |
| Deletion | Chr2 | 4493127 | 4493128 | 1 | |
| Deletion | Chr2 | 4973206 | 4973216 | 10 | |
| Deletion | Chr9 | 7759692 | 7759700 | 8 | |
| Deletion | Chr8 | 9184146 | 9184149 | 3 | |
| Deletion | Chr10 | 17560534 | 17560536 | 2 | |
| Deletion | Chr7 | 19385685 | 19385690 | 5 | |
| Deletion | Chr6 | 20418660 | 20418669 | 9 | |
| Deletion | Chr8 | 20766575 | 20766577 | 2 | |
| Deletion | Chr6 | 24234576 | 24234579 | 3 | LOC_Os06g40650 |
| Deletion | Chr8 | 24679756 | 24679758 | 2 | LOC_Os08g39060 |
| Deletion | Chr7 | 24718579 | 24718583 | 4 | |
| Deletion | Chr6 | 25320911 | 25320919 | 8 | |
| Deletion | Chr6 | 25395001 | 25452000 | 57000 | 8 |
| Deletion | Chr1 | 28534982 | 28534983 | 1 | LOC_Os01g49630 |
| Deletion | Chr1 | 30005111 | 30005112 | 1 | |
| Deletion | Chr2 | 31817436 | 31817457 | 21 | |
| Deletion | Chr2 | 32796414 | 32796417 | 3 | |
| Deletion | Chr4 | 34248434 | 34248435 | 1 | |
| Deletion | Chr3 | 34508301 | 34508316 | 15 | LOC_Os03g60720 |
| Deletion | Chr4 | 34517057 | 34517061 | 4 | LOC_Os04g57940 |
| Deletion | Chr1 | 34902045 | 34902063 | 18 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 10958316 | 10958316 | 1 | |
| Insertion | Chr7 | 28111659 | 28111659 | 1 | |
| Insertion | Chr8 | 131623 | 131623 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 14658560 | 14709290 | LOC_Os08g24370 |
| Inversion | Chr7 | 18399408 | 18809404 | |
| Inversion | Chr6 | 25378040 | 25388712 | LOC_Os06g42270 |
| Inversion | Chr2 | 27656939 | 30574488 | LOC_Os02g50040 |
| Inversion | Chr2 | 27730899 | 30574486 | LOC_Os02g50040 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 2634626 | Chr7 | 24345706 | LOC_Os07g40620 |
| Translocation | Chr11 | 2634645 | Chr7 | 24346004 | |
| Translocation | Chr10 | 9167693 | Chr1 | 2332983 | LOC_Os10g18080 |
| Translocation | Chr10 | 9167699 | Chr1 | 2330205 | LOC_Os10g18080 |
| Translocation | Chr8 | 14658591 | Chr7 | 14162997 | LOC_Os07g24910 |
| Translocation | Chr8 | 14709295 | Chr7 | 14166407 | LOC_Os08g24370 |
| Translocation | Chr11 | 18032334 | Chr5 | 14982105 | LOC_Os05g25750 |
