Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN685-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN685-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 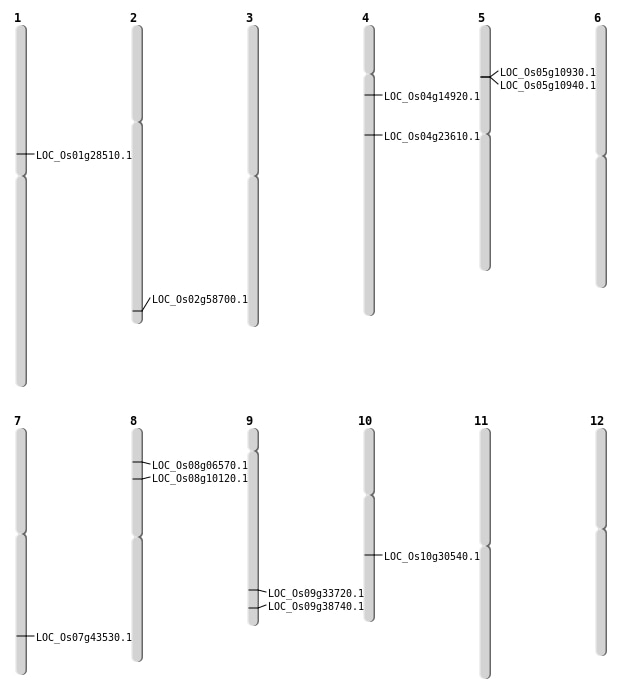
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 41329004 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 15877862 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30540 |
| SBS | Chr10 | 18066595 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 18066596 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 9357337 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 15534128 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 21869471 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 7794657 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 29018066 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 31406358 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 19272689 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 21883383 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 3002231 | G-C | HET | INTRON | |
| SBS | Chr4 | 5206290 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 8393279 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g14920 |
| SBS | Chr6 | 1915225 | G-A | HET | INTRON | |
| SBS | Chr6 | 1915226 | C-A | HET | INTRON | |
| SBS | Chr7 | 20923497 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 24530253 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 1791463 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 18620078 | C-T | HET | INTRON | |
| SBS | Chr8 | 3709067 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g06570 |
| SBS | Chr8 | 3930624 | G-A | HET | INTRON | |
| SBS | Chr8 | 5873310 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g10120 |
| SBS | Chr9 | 21294720 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 22264156 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g38740 |
| SBS | Chr9 | 22264161 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 2928248 | A-T | HET | INTERGENIC |
Deletions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 6070001 | 6095000 | 25000 | 2 |
| Deletion | Chr4 | 11897878 | 11897885 | 7 | |
| Deletion | Chr3 | 12283646 | 12283656 | 10 | |
| Deletion | Chr4 | 13519808 | 13519810 | 2 | LOC_Os04g23610 |
| Deletion | Chr3 | 15555320 | 15555322 | 2 | |
| Deletion | Chr3 | 17488493 | 17488494 | 1 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 11351829 | 11351830 | 2 | |
| Insertion | Chr5 | 28034317 | 28034317 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 6070103 | 7308057 | |
| Inversion | Chr5 | 6094866 | 7308061 | |
| Inversion | Chr1 | 15961372 | 15966406 | LOC_Os01g28510 |
| Inversion | Chr7 | 26041244 | 28311667 | LOC_Os07g43530 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 547818 | Chr2 | 35865577 | LOC_Os02g58700 |
| Translocation | Chr5 | 547840 | Chr2 | 35865588 | LOC_Os02g58700 |
| Translocation | Chr9 | 19918570 | Chr7 | 26319114 | LOC_Os09g33720 |
