Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN725-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN725-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 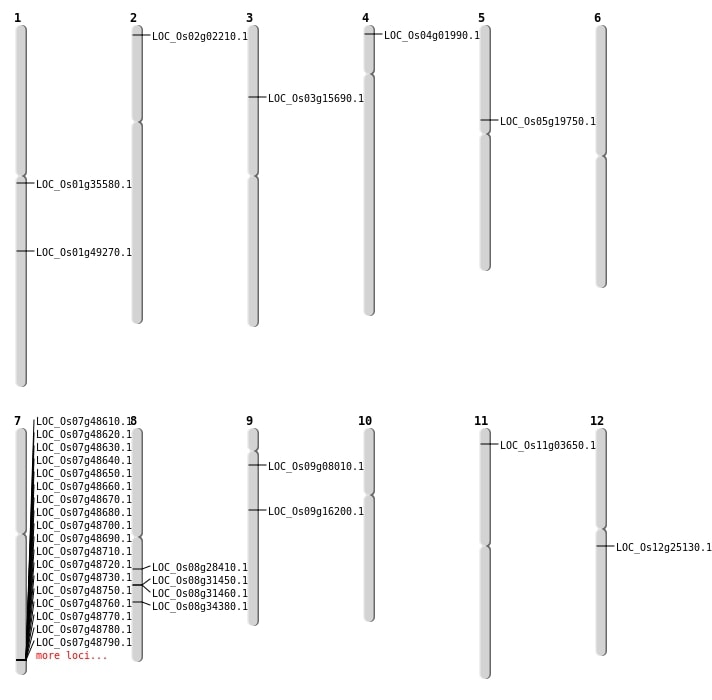
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11397015 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 39059943 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 41675745 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 1420407 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g03650 |
| SBS | Chr11 | 778173 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 14433438 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr12 | 16283391 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 24011281 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 25724354 | A-G | HET | INTRON | |
| SBS | Chr2 | 10161672 | C-A | HET | INTRON | |
| SBS | Chr2 | 12627403 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 27272494 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 13148472 | A-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 15737789 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 10689738 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 10581501 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 11528309 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g19750 |
| SBS | Chr5 | 12122345 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 4703009 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 4898749 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 13873511 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 22044214 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 26011967 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 28530643 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 28530645 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 6136143 | C-T | HET | INTRON | |
| SBS | Chr7 | 22292432 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 18808807 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 19590024 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 9191549 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 4125477 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g08010 |
| SBS | Chr9 | 9882035 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g16200 |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 24557 | 24558 | 1 | |
| Deletion | Chr4 | 614802 | 614803 | 1 | LOC_Os04g01990 |
| Deletion | Chr2 | 695673 | 701533 | 5860 | LOC_Os02g02210 |
| Deletion | Chr9 | 2273382 | 2273490 | 108 | |
| Deletion | Chr4 | 2307883 | 2307886 | 3 | |
| Deletion | Chr9 | 3055434 | 3055440 | 6 | |
| Deletion | Chr5 | 4765180 | 4765183 | 3 | |
| Deletion | Chr3 | 4820671 | 4820672 | 1 | |
| Deletion | Chr5 | 4912503 | 4912536 | 33 | |
| Deletion | Chr12 | 5471658 | 5471662 | 4 | |
| Deletion | Chr4 | 5901263 | 5901268 | 5 | |
| Deletion | Chr9 | 8354968 | 8354972 | 4 | |
| Deletion | Chr7 | 8388856 | 8388858 | 2 | |
| Deletion | Chr3 | 8656516 | 8656518 | 2 | LOC_Os03g15690 |
| Deletion | Chr12 | 10913293 | 10913294 | 1 | |
| Deletion | Chr12 | 14433338 | 14433351 | 13 | LOC_Os12g25130 |
| Deletion | Chr10 | 14722838 | 14722843 | 5 | |
| Deletion | Chr7 | 15703362 | 15703373 | 11 | |
| Deletion | Chr8 | 17329001 | 17344000 | 15000 | LOC_Os08g28410 |
| Deletion | Chr5 | 18583100 | 18583108 | 8 | |
| Deletion | Chr8 | 19448001 | 19462000 | 14000 | 2 |
| Deletion | Chr8 | 21317720 | 21317731 | 11 | |
| Deletion | Chr8 | 21581698 | 21581702 | 4 | LOC_Os08g34380 |
| Deletion | Chr12 | 21670618 | 21670634 | 16 | |
| Deletion | Chr4 | 21979328 | 21979336 | 8 | |
| Deletion | Chr11 | 23393339 | 23393340 | 1 | |
| Deletion | Chr1 | 28324951 | 28325104 | 153 | LOC_Os01g49270 |
| Deletion | Chr7 | 29102514 | 29347325 | 244811 | 38 |
Insertions: 14
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 1800464 | 16822216 | |
| Inversion | Chr8 | 17260253 | 17329324 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 4022264 | Chr1 | 19682119 | LOC_Os01g35580 |
| Translocation | Chr12 | 17012452 | Chr9 | 2273529 | |
| Translocation | Chr12 | 17012609 | Chr9 | 2273383 | |
| Translocation | Chr6 | 21999787 | Chr2 | 15411038 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr8 | 17260259 | 17453750 | 193491 |
