Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN731-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN731-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 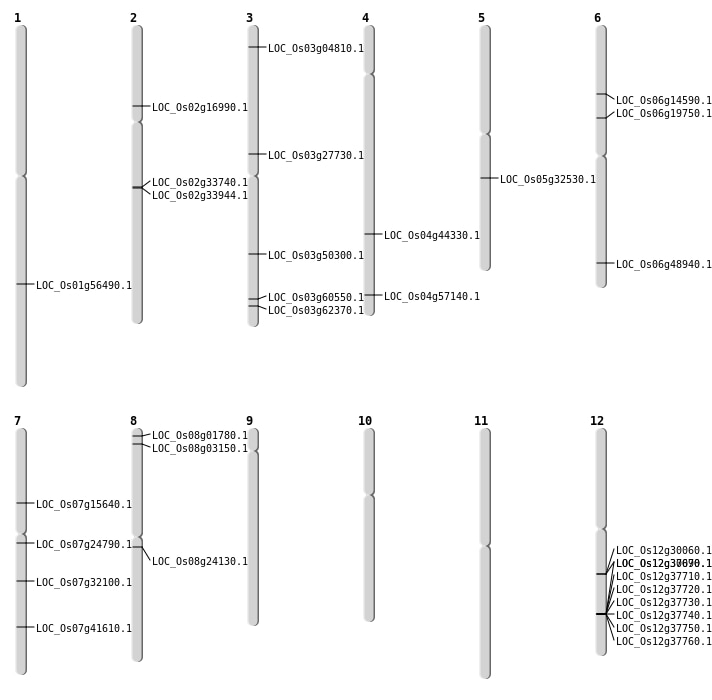
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 29482990 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 15173895 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 3176816 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 7881643 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 19883002 | T-A | HOMO | INTRON | |
| SBS | Chr2 | 19883003 | T-A | HOMO | INTRON | |
| SBS | Chr2 | 19883005 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 19883448 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 2287852 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g04810 |
| SBS | Chr3 | 26754331 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 28165344 | A-G | HET | INTRON | |
| SBS | Chr3 | 31067921 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 4551699 | G-A | HET | INTRON | |
| SBS | Chr4 | 13696780 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 14607170 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 26447484 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2693238 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 27942325 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 33598877 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 12669415 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 14631535 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 16917794 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 26741007 | C-T | HET | INTRON | |
| SBS | Chr6 | 26817035 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 8212931 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g14590 |
| SBS | Chr7 | 19074179 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g32100 |
| SBS | Chr8 | 12895314 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 1448792 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g03150 |
| SBS | Chr8 | 14572099 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g24130 |
| SBS | Chr8 | 5194750 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 22752086 | A-T | HET | UTR_3_PRIME |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2400866 | 2400883 | 17 | |
| Deletion | Chr5 | 2730829 | 2730830 | 1 | |
| Deletion | Chr12 | 3018161 | 3018169 | 8 | |
| Deletion | Chr1 | 3377845 | 3377851 | 6 | |
| Deletion | Chr7 | 9076013 | 9076020 | 7 | LOC_Os07g15640 |
| Deletion | Chr2 | 9700150 | 9700165 | 15 | LOC_Os02g16990 |
| Deletion | Chr8 | 10695491 | 10695499 | 8 | |
| Deletion | Chr6 | 11288582 | 11288584 | 2 | LOC_Os06g19750 |
| Deletion | Chr7 | 14092810 | 14092858 | 48 | LOC_Os07g24790 |
| Deletion | Chr11 | 14992007 | 14992008 | 1 | |
| Deletion | Chr3 | 15894063 | 15894065 | 2 | LOC_Os03g27730 |
| Deletion | Chr12 | 18003001 | 18010000 | 7000 | 2 |
| Deletion | Chr5 | 19018995 | 19018997 | 2 | LOC_Os05g32530 |
| Deletion | Chr4 | 20619099 | 20619101 | 2 | |
| Deletion | Chr9 | 21409215 | 21409217 | 2 | |
| Deletion | Chr12 | 23134001 | 23186000 | 52000 | 7 |
| Deletion | Chr5 | 25926630 | 25926631 | 1 | |
| Deletion | Chr4 | 26256891 | 26256892 | 1 | LOC_Os04g44330 |
| Deletion | Chr3 | 28672139 | 28672148 | 9 | LOC_Os03g50300 |
| Deletion | Chr7 | 29381479 | 29381732 | 253 | |
| Deletion | Chr6 | 29629238 | 29629251 | 13 | LOC_Os06g48940 |
| Deletion | Chr1 | 32552971 | 32552984 | 13 | LOC_Os01g56490 |
| Deletion | Chr4 | 34045297 | 34045307 | 10 | LOC_Os04g57140 |
| Deletion | Chr3 | 34212755 | 34212756 | 1 | |
| Deletion | Chr3 | 35332739 | 35332778 | 39 | LOC_Os03g62370 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 6720241 | 6720242 | 2 | |
| Insertion | Chr2 | 26188425 | 26188426 | 2 | |
| Insertion | Chr2 | 9397776 | 9397776 | 1 | |
| Insertion | Chr3 | 10285971 | 10285971 | 1 |
No Inversion
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 470516 | Chr1 | 25151223 | LOC_Os08g01780 |
| Translocation | Chr9 | 2717573 | Chr7 | 24948166 | LOC_Os07g41610 |
| Translocation | Chr5 | 3174705 | Chr1 | 25151227 | |
| Translocation | Chr5 | 3894062 | Chr3 | 34416788 | LOC_Os03g60550 |
| Translocation | Chr4 | 16367931 | Chr3 | 20800548 | |
| Translocation | Chr12 | 18010197 | Chr8 | 470523 | 2 |
| Translocation | Chr12 | 21229841 | Chr4 | 16367939 | |
| Translocation | Chr12 | 23186076 | Chr2 | 20131910 | 2 |
| Translocation | Chr12 | 23189598 | Chr2 | 20230653 | LOC_Os02g33944 |
