Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN765-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN765-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 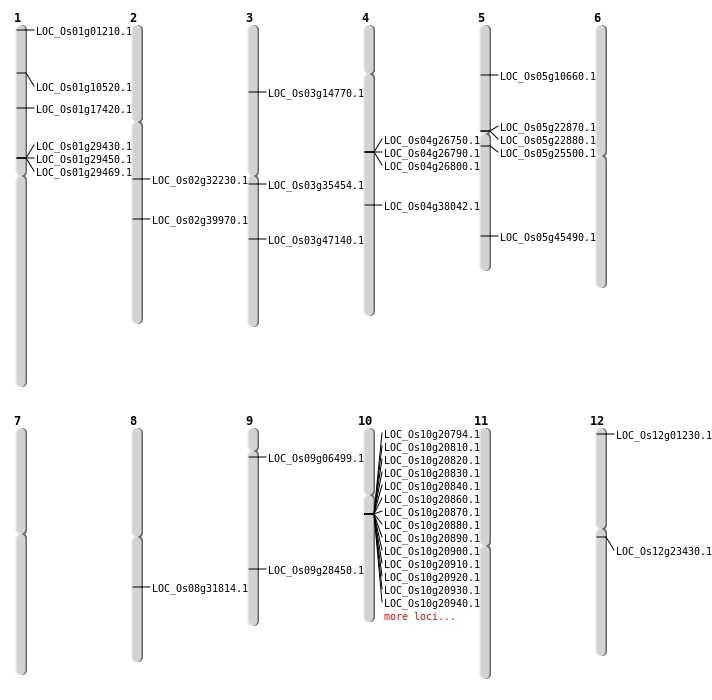
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10020327 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g17420 |
| SBS | Chr1 | 24190957 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 37317282 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 99644 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g01210 |
| SBS | Chr10 | 134377 | C-A | HET | INTRON | |
| SBS | Chr11 | 21640306 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 3943747 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 21762658 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 24157911 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 13976582 | G-C | HET | INTRON | |
| SBS | Chr3 | 15761652 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 8030231 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g14770 |
| SBS | Chr4 | 1664330 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 4595511 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 5758802 | A-C | HOMO | INTRON | |
| SBS | Chr5 | 6942827 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 8414287 | C-T | HET | INTRON | |
| SBS | Chr6 | 3444421 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 5828084 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 8173736 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 8173737 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 16675960 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 27278736 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 19811355 | G-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 14641522 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 1546949 | C-T | HET | INTRON | |
| SBS | Chr9 | 17312171 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g28450 |
| SBS | Chr9 | 17657232 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 17657233 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 4147111 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 5165371 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 9466377 | G-C | HET | SYNONYMOUS_CODING |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 135809 | 135816 | 7 | LOC_Os12g01230 |
| Deletion | Chr10 | 278695 | 278696 | 1 | |
| Deletion | Chr9 | 3082788 | 3083326 | 538 | |
| Deletion | Chr1 | 5585031 | 5585037 | 6 | LOC_Os01g10520 |
| Deletion | Chr5 | 5834092 | 5834102 | 10 | LOC_Os05g10660 |
| Deletion | Chr5 | 8985143 | 8985151 | 8 | |
| Deletion | Chr10 | 10548001 | 10790000 | 242000 | 36 |
| Deletion | Chr1 | 11371371 | 11371372 | 1 | |
| Deletion | Chr9 | 12834521 | 12834522 | 1 | |
| Deletion | Chr5 | 12986001 | 13001000 | 15000 | 2 |
| Deletion | Chr12 | 14596735 | 14596740 | 5 | |
| Deletion | Chr5 | 14825559 | 14825564 | 5 | LOC_Os05g25500 |
| Deletion | Chr4 | 15627001 | 15684000 | 57000 | 2 |
| Deletion | Chr4 | 15789001 | 15821000 | 32000 | LOC_Os04g26800 |
| Deletion | Chr1 | 16496767 | 16513044 | 16277 | 3 |
| Deletion | Chr9 | 17280411 | 17280435 | 24 | |
| Deletion | Chr2 | 19039414 | 19039423 | 9 | LOC_Os02g32230 |
| Deletion | Chr8 | 19201144 | 19201157 | 13 | |
| Deletion | Chr3 | 19656362 | 19656455 | 93 | LOC_Os03g35454 |
| Deletion | Chr4 | 22646989 | 22646996 | 7 | LOC_Os04g38042 |
| Deletion | Chr2 | 24169075 | 24169076 | 1 | LOC_Os02g39970 |
| Deletion | Chr6 | 24545892 | 24545912 | 20 | |
| Deletion | Chr7 | 27652760 | 27652799 | 39 | |
| Deletion | Chr3 | 28786235 | 28786238 | 3 | |
| Deletion | Chr2 | 29394461 | 29394463 | 2 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 40095816 | 40095816 | 1 | |
| Insertion | Chr11 | 28676529 | 28676531 | 3 | |
| Insertion | Chr5 | 26384202 | 26384202 | 1 | LOC_Os05g45490 |
| Insertion | Chr6 | 30396307 | 30396308 | 2 |
No Inversion
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 3082796 | Chr3 | 26679200 | 2 |
| Translocation | Chr9 | 3083317 | Chr3 | 26679193 | 2 |
| Translocation | Chr12 | 13246863 | Chr1 | 5488390 | LOC_Os12g23430 |
| Translocation | Chr8 | 19732245 | Chr4 | 16243641 | LOC_Os08g31814 |
| Translocation | Chr8 | 19732248 | Chr4 | 16241022 | LOC_Os08g31814 |
