Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN770-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN770-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 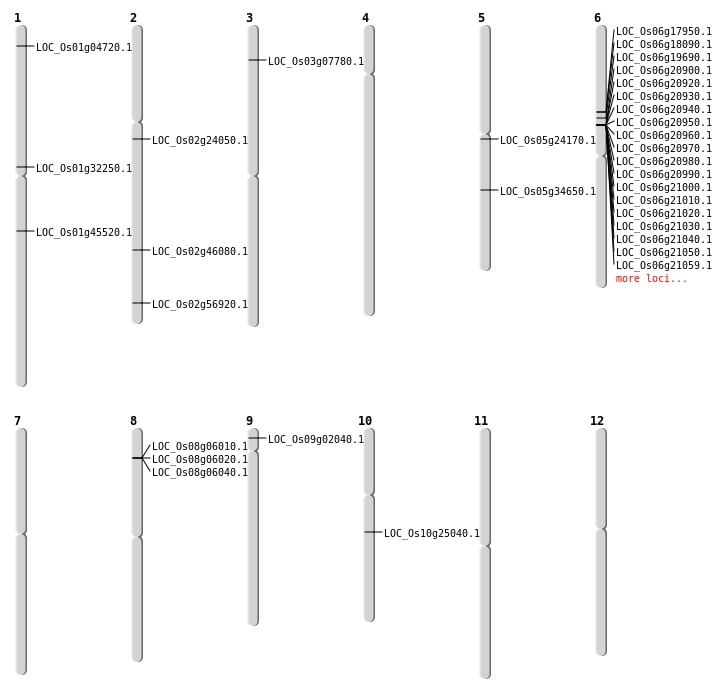
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2058073 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 20752690 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 21079068 | C-A | HOMO | INTRON | |
| SBS | Chr1 | 25839620 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 25839636 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g45520 |
| SBS | Chr1 | 27213089 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 29990658 | G-A | HET | INTRON | |
| SBS | Chr1 | 39823700 | G-A | HOMO | UTR_5_PRIME | |
| SBS | Chr10 | 17848144 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 7391556 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 16886459 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 16994694 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 13938169 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g24050 |
| SBS | Chr3 | 1283290 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 18174266 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 23590116 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 33122697 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 33230814 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 3335195 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 3952952 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g07780 |
| SBS | Chr3 | 8847488 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 13402170 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 16515706 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 6628407 | G-A | HET | INTRON | |
| SBS | Chr5 | 12935844 | T-A | HOMO | INTRON | |
| SBS | Chr5 | 13682218 | C-A | HET | INTRON | |
| SBS | Chr5 | 20551739 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g34650 |
| SBS | Chr5 | 843639 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 14358672 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 29685794 | C-T | HET | INTRON | |
| SBS | Chr7 | 28171067 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 3804896 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 3031866 | G-A | HET | INTRON | |
| SBS | Chr9 | 8388726 | C-G | HET | SYNONYMOUS_CODING |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 742516 | 742717 | 201 | |
| Deletion | Chr12 | 2127317 | 2127318 | 1 | |
| Deletion | Chr1 | 2134246 | 2134251 | 5 | LOC_Os01g04720 |
| Deletion | Chr8 | 3272001 | 3298000 | 26000 | 3 |
| Deletion | Chr8 | 3274920 | 3298242 | 23322 | 3 |
| Deletion | Chr6 | 3748576 | 3748586 | 10 | |
| Deletion | Chr4 | 4249395 | 4249405 | 10 | |
| Deletion | Chr6 | 4787933 | 4787934 | 1 | |
| Deletion | Chr2 | 5400215 | 5400217 | 2 | |
| Deletion | Chr8 | 7384957 | 7384974 | 17 | |
| Deletion | Chr2 | 8095303 | 8104649 | 9346 | |
| Deletion | Chr5 | 8904939 | 8905057 | 118 | |
| Deletion | Chr2 | 10363081 | 10363082 | 1 | |
| Deletion | Chr6 | 12076712 | 12264941 | 188229 | 32 |
| Deletion | Chr11 | 12077270 | 12077375 | 105 | |
| Deletion | Chr5 | 12360953 | 12360960 | 7 | |
| Deletion | Chr12 | 12363298 | 12363300 | 2 | |
| Deletion | Chr10 | 12892387 | 12892398 | 11 | LOC_Os10g25040 |
| Deletion | Chr5 | 12999708 | 12999709 | 1 | |
| Deletion | Chr5 | 12999712 | 12999713 | 1 | |
| Deletion | Chr5 | 13708086 | 13708095 | 9 | |
| Deletion | Chr5 | 13954452 | 13954463 | 11 | LOC_Os05g24170 |
| Deletion | Chr2 | 15240483 | 15240485 | 2 | |
| Deletion | Chr3 | 15337217 | 15337268 | 51 | |
| Deletion | Chr2 | 16984490 | 16984502 | 12 | |
| Deletion | Chr1 | 17091157 | 17091168 | 11 | |
| Deletion | Chr1 | 17667411 | 17667412 | 1 | LOC_Os01g32250 |
| Deletion | Chr10 | 18899571 | 18899572 | 1 | |
| Deletion | Chr1 | 19085239 | 19085263 | 24 | |
| Deletion | Chr11 | 23493161 | 23493169 | 8 | |
| Deletion | Chr5 | 28160386 | 28160390 | 4 | |
| Deletion | Chr6 | 28386616 | 28386617 | 1 | |
| Deletion | Chr6 | 28386620 | 28386621 | 1 | |
| Deletion | Chr4 | 31409347 | 31409353 | 6 | |
| Deletion | Chr2 | 34889694 | 34889697 | 3 | LOC_Os02g56920 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 28081723 | 28081724 | 2 | LOC_Os02g46080 |
| Insertion | Chr7 | 15284994 | 15284995 | 2 | |
| Insertion | Chr8 | 16622730 | 16622731 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 742521 | 17915185 | LOC_Os09g02040 |
| Inversion | Chr9 | 742714 | 17915182 | LOC_Os09g02040 |
| Inversion | Chr6 | 10435868 | 10535179 | 2 |
| Inversion | Chr6 | 10440691 | 11665287 | LOC_Os06g17950 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 11248205 | Chr3 | 3336000 | LOC_Os06g19690 |
