Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN780-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN780-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 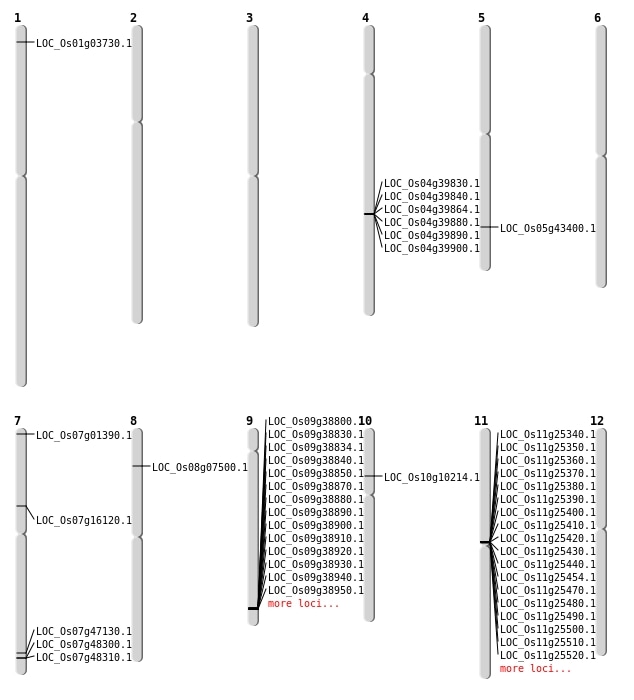
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1568109 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g03730 |
| SBS | Chr1 | 5267825 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 10556974 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 13027649 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 4081801 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 5619500 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g10214 |
| SBS | Chr10 | 8537483 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 13137422 | C-A | HET | INTRON | |
| SBS | Chr11 | 14605784 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g25610 |
| SBS | Chr11 | 17588804 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g30260 |
| SBS | Chr11 | 27178687 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 5299755 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 8115628 | T-A | HOMO | INTRON | |
| SBS | Chr12 | 17528563 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 18269558 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 18911086 | A-T | HET | INTRON | |
| SBS | Chr12 | 24018585 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 24242495 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 1445012 | A-G | HET | INTRON | |
| SBS | Chr2 | 1901061 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 3484076 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 35595145 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 9863238 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 25236122 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g43400 |
| SBS | Chr6 | 15066808 | A-G | HET | INTRON | |
| SBS | Chr6 | 22523595 | T-A | HET | INTRON | |
| SBS | Chr6 | 30125245 | T-G | HET | INTRON | |
| SBS | Chr6 | 3171910 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 3373274 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 3699470 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 407264 | T-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 1065828 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 243316 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g01390 |
| SBS | Chr7 | 2656638 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 9387377 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16120 |
| SBS | Chr8 | 22283208 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 3948926 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 12981996 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 5400444 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5400445 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 737526 | C-T | HET | SYNONYMOUS_CODING |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 4005759 | 4005766 | 7 | |
| Deletion | Chr8 | 4212658 | 4212659 | 1 | LOC_Os08g07500 |
| Deletion | Chr9 | 5194815 | 5194826 | 11 | |
| Deletion | Chr10 | 5607782 | 5607783 | 1 | |
| Deletion | Chr4 | 7664588 | 7664591 | 3 | |
| Deletion | Chr2 | 7927694 | 7927695 | 1 | |
| Deletion | Chr9 | 9485730 | 9485737 | 7 | |
| Deletion | Chr1 | 9565715 | 9565716 | 1 | |
| Deletion | Chr7 | 9989850 | 9989851 | 1 | |
| Deletion | Chr2 | 11601457 | 11601480 | 23 | |
| Deletion | Chr9 | 12228609 | 12228611 | 2 | |
| Deletion | Chr8 | 14177312 | 14177322 | 10 | |
| Deletion | Chr11 | 14442001 | 15121000 | 679000 | 97 |
| Deletion | Chr12 | 14452634 | 14452646 | 12 | |
| Deletion | Chr3 | 17869113 | 17869115 | 2 | |
| Deletion | Chr4 | 18799652 | 18799662 | 10 | |
| Deletion | Chr5 | 21968429 | 21968433 | 4 | |
| Deletion | Chr9 | 22293001 | 22334000 | 41000 | 8 |
| Deletion | Chr9 | 22342001 | 22623000 | 281000 | 42 |
| Deletion | Chr4 | 23724001 | 23770000 | 46000 | 6 |
| Deletion | Chr7 | 28182548 | 28182550 | 2 | LOC_Os07g47130 |
| Deletion | Chr7 | 28862112 | 28864192 | 2080 | 2 |
| Deletion | Chr3 | 32870536 | 32870570 | 34 | |
| Deletion | Chr1 | 41181382 | 41181390 | 8 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 9425088 | 9425105 | 18 | |
| Insertion | Chr11 | 28281687 | 28281690 | 4 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 3788354 | 3842672 | |
| Inversion | Chr8 | 3842683 | 5699007 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 22623633 | Chr7 | 28862126 | 2 |
