Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN786-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN786-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 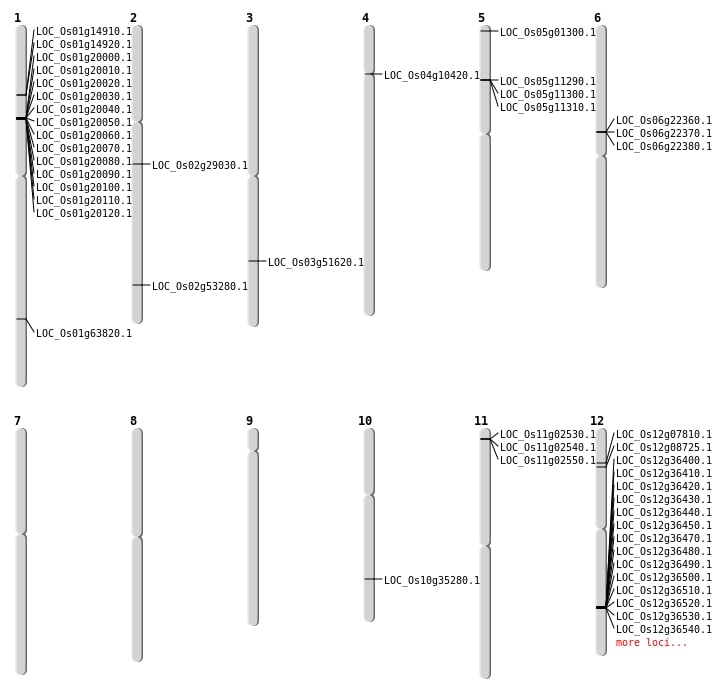
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15862234 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 19493616 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 20456357 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 8324780 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 17109527 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 18852446 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g35280 |
| SBS | Chr11 | 11308733 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 11974037 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 27304108 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 7392568 | C-T | HET | INTRON | |
| SBS | Chr12 | 26423959 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 10867285 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 11535329 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 21064027 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 23607059 | A-C | HET | UTR_5_PRIME | |
| SBS | Chr2 | 34768141 | G-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 11830795 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 20299021 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 4761374 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 6350233 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 10918245 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 16378789 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 29257552 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 29257553 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 6336268 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 6784299 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 8117495 | C-A | HET | INTRON | |
| SBS | Chr6 | 17115600 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 30435943 | C-T | HET | INTRON | |
| SBS | Chr7 | 12298513 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14901129 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr7 | 22027589 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 18391490 | G-C | HOMO | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 196147 | 196153 | 6 | LOC_Os05g01300 |
| Deletion | Chr4 | 463490 | 463492 | 2 | |
| Deletion | Chr11 | 788060 | 804414 | 16354 | 3 |
| Deletion | Chr8 | 1664924 | 1664936 | 12 | |
| Deletion | Chr2 | 3773161 | 3773194 | 33 | |
| Deletion | Chr12 | 3947113 | 3947123 | 10 | LOC_Os12g07810 |
| Deletion | Chr12 | 4459637 | 4459648 | 11 | LOC_Os12g08725 |
| Deletion | Chr4 | 5661877 | 5661908 | 31 | LOC_Os04g10420 |
| Deletion | Chr5 | 6372001 | 6390000 | 18000 | 3 |
| Deletion | Chr11 | 7315717 | 7315721 | 4 | |
| Deletion | Chr1 | 8352001 | 8362000 | 10000 | 2 |
| Deletion | Chr11 | 8966349 | 8966360 | 11 | |
| Deletion | Chr9 | 9186829 | 9186833 | 4 | |
| Deletion | Chr10 | 11242403 | 11242411 | 8 | |
| Deletion | Chr1 | 11366579 | 11435933 | 69354 | 13 |
| Deletion | Chr6 | 12980626 | 12996734 | 16108 | 3 |
| Deletion | Chr2 | 13562822 | 13562852 | 30 | |
| Deletion | Chr2 | 14618912 | 14619492 | 580 | |
| Deletion | Chr8 | 20020148 | 20020265 | 117 | |
| Deletion | Chr3 | 20880338 | 20880346 | 8 | |
| Deletion | Chr12 | 22283000 | 23141538 | 858538 | 130 |
| Deletion | Chr2 | 22986771 | 22986778 | 7 | |
| Deletion | Chr11 | 26413963 | 26413966 | 3 | |
| Deletion | Chr7 | 27801052 | 27801097 | 45 | |
| Deletion | Chr5 | 28776362 | 28776366 | 4 | |
| Deletion | Chr5 | 29363164 | 29363171 | 7 | |
| Deletion | Chr3 | 29792725 | 29792737 | 12 | |
| Deletion | Chr2 | 32613286 | 32613287 | 1 | LOC_Os02g53280 |
| Deletion | Chr1 | 37026858 | 37026879 | 21 | LOC_Os01g63820 |
Insertions: 7
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 6371215 | 6407170 | |
| Inversion | Chr5 | 6389828 | 6408146 | |
| Inversion | Chr5 | 6407185 | 12999895 | |
| Inversion | Chr5 | 6408142 | 12999882 | |
| Inversion | Chr2 | 12077386 | 17190738 | LOC_Os02g29030 |
| Inversion | Chr12 | 22283015 | 23197212 | LOC_Os12g37780 |
| Inversion | Chr12 | 23141536 | 23197080 | LOC_Os12g37780 |
| Inversion | Chr1 | 30299293 | 30470106 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 29549601 | Chr2 | 14618916 | LOC_Os03g51620 |
