Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN788-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN788-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 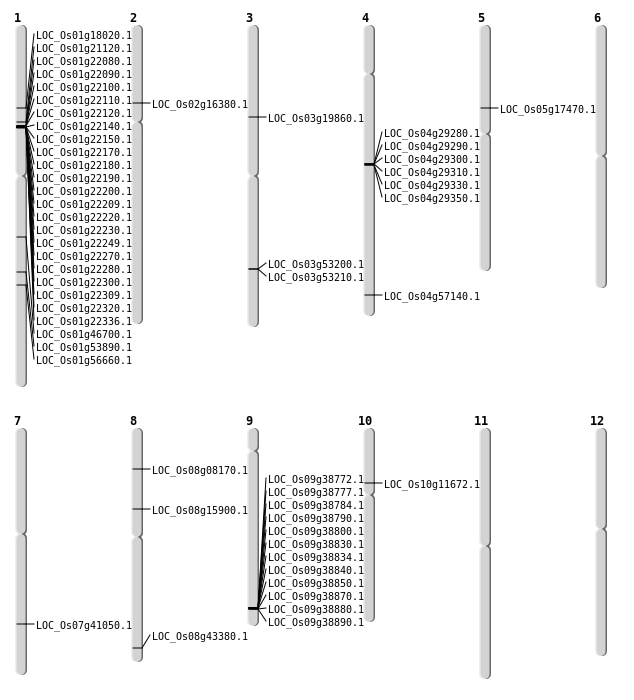
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10082103 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g18020 |
| SBS | Chr1 | 10849895 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 14170742 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 19155951 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 25848513 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 26592249 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 30985422 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g53890 |
| SBS | Chr1 | 32696275 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g56660 |
| SBS | Chr1 | 34815254 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 582253 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 6443892 | A-T | HET | STOP_GAINED | LOC_Os10g11672 |
| SBS | Chr11 | 27142444 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 27142445 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 9949004 | A-G | HOMO | INTRON | |
| SBS | Chr12 | 18011404 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 4334185 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 15676946 | G-A | HET | INTRON | |
| SBS | Chr2 | 19221150 | A-G | HET | INTRON | |
| SBS | Chr2 | 3959736 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 4367213 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 6056748 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 9333481 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g16380 |
| SBS | Chr3 | 11171011 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g19860 |
| SBS | Chr3 | 21273927 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 4254068 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 10037400 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17470 |
| SBS | Chr5 | 22697776 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 29053005 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 29390775 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 8142875 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 25436219 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 9266511 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 22606355 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 24555291 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g41050 |
| SBS | Chr7 | 27502423 | G-A | HET | INTRON | |
| SBS | Chr8 | 1001007 | G-A | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1362113 | 1362140 | 27 | |
| Deletion | Chr8 | 4651511 | 4651514 | 3 | LOC_Os08g08170 |
| Deletion | Chr3 | 4827976 | 4828027 | 51 | |
| Deletion | Chr5 | 5266922 | 5266923 | 1 | |
| Deletion | Chr8 | 9688690 | 9688693 | 3 | LOC_Os08g15900 |
| Deletion | Chr3 | 10479447 | 10479453 | 6 | |
| Deletion | Chr6 | 12180778 | 12180779 | 1 | |
| Deletion | Chr1 | 12401001 | 12559000 | 158000 | 21 |
| Deletion | Chr12 | 13032021 | 13032022 | 1 | |
| Deletion | Chr7 | 16638416 | 16638422 | 6 | |
| Deletion | Chr4 | 17363001 | 17432000 | 69000 | 6 |
| Deletion | Chr11 | 21251592 | 21251606 | 14 | |
| Deletion | Chr9 | 22278001 | 22334000 | 56000 | 12 |
| Deletion | Chr5 | 22686032 | 22686037 | 5 | |
| Deletion | Chr1 | 25085242 | 25085255 | 13 | |
| Deletion | Chr1 | 26592027 | 26592028 | 1 | LOC_Os01g46700 |
| Deletion | Chr12 | 26788904 | 26788905 | 1 | |
| Deletion | Chr8 | 27410379 | 27410380 | 1 | LOC_Os08g43380 |
| Deletion | Chr8 | 28361044 | 28361045 | 1 | |
| Deletion | Chr3 | 28654999 | 28655029 | 30 | |
| Deletion | Chr3 | 30520244 | 30520271 | 27 | 2 |
| Deletion | Chr3 | 31839400 | 31839410 | 10 | |
| Deletion | Chr4 | 34048829 | 34048830 | 1 | LOC_Os04g57140 |
Insertions: 7
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 11284570 | 11784293 | LOC_Os01g21120 |
| Inversion | Chr1 | 11284592 | 11784295 | LOC_Os01g21120 |
No Translocation
