Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN796-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN796-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 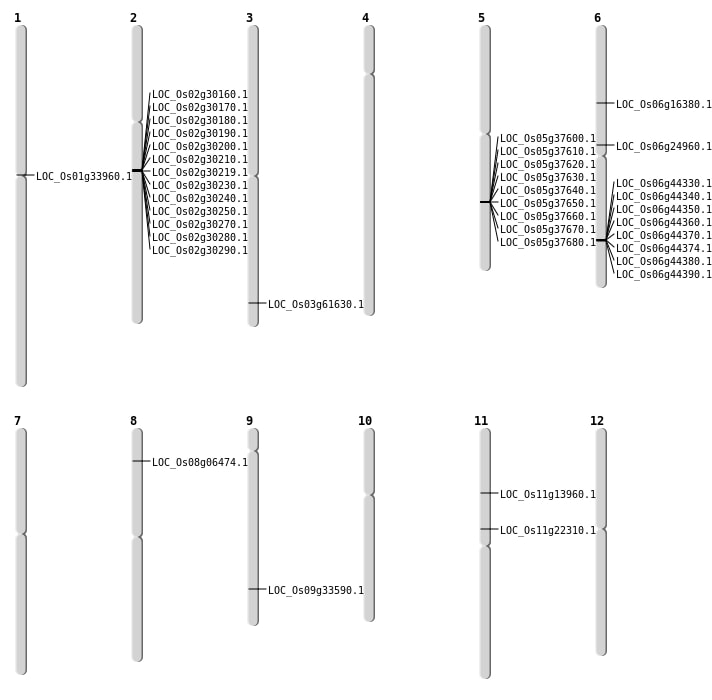
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26760468 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 5077408 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 12206886 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 172771 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 2767119 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 12788717 | G-A | HET | STOP_GAINED | LOC_Os11g22310 |
| SBS | Chr11 | 7726348 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g13960 |
| SBS | Chr12 | 13813340 | C-T | HET | INTRON | |
| SBS | Chr12 | 22198542 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 27207326 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 22415819 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 2867217 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 30277452 | G-T | HOMO | INTRON | |
| SBS | Chr3 | 22301972 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 22301974 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 26796127 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 29733628 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 16440737 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 19950274 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 26258120 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr4 | 28034425 | C-A | HET | INTRON | |
| SBS | Chr4 | 30091908 | T-A | HET | INTRON | |
| SBS | Chr5 | 14607903 | T-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 17919389 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 13525600 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 10483236 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 25445365 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 25445368 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 28469355 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 6894587 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 21109574 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 3636888 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g06474 |
| SBS | Chr9 | 19820852 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g33590 |
| SBS | Chr9 | 7083411 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 7259483 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 956648 | T-G | HET | INTERGENIC |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2774700 | 2774708 | 8 | |
| Deletion | Chr3 | 6906298 | 6906300 | 2 | |
| Deletion | Chr3 | 6963264 | 6963282 | 18 | |
| Deletion | Chr5 | 14607888 | 14607901 | 13 | |
| Deletion | Chr2 | 16200207 | 16200212 | 5 | |
| Deletion | Chr2 | 17918001 | 18013000 | 95000 | 13 |
| Deletion | Chr5 | 18443973 | 18443974 | 1 | |
| Deletion | Chr2 | 19404940 | 19404948 | 8 | |
| Deletion | Chr5 | 22007001 | 22058000 | 51000 | 9 |
| Deletion | Chr9 | 22529982 | 22529983 | 1 | |
| Deletion | Chr6 | 26759001 | 26803000 | 44000 | 8 |
| Deletion | Chr2 | 28850849 | 28850851 | 2 | |
| Deletion | Chr1 | 30257717 | 30257718 | 1 | |
| Deletion | Chr3 | 34940961 | 34940968 | 7 | LOC_Os03g61630 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 21795985 | 21795986 | 2 | |
| Insertion | Chr8 | 17037281 | 17037282 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 20443956 | 20549119 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 9350367 | Chr1 | 18685174 | 2 |
| Translocation | Chr6 | 14623557 | Chr2 | 18013290 | 2 |
| Translocation | Chr7 | 28618359 | Chr1 | 25847638 |
