Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN804-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN804-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 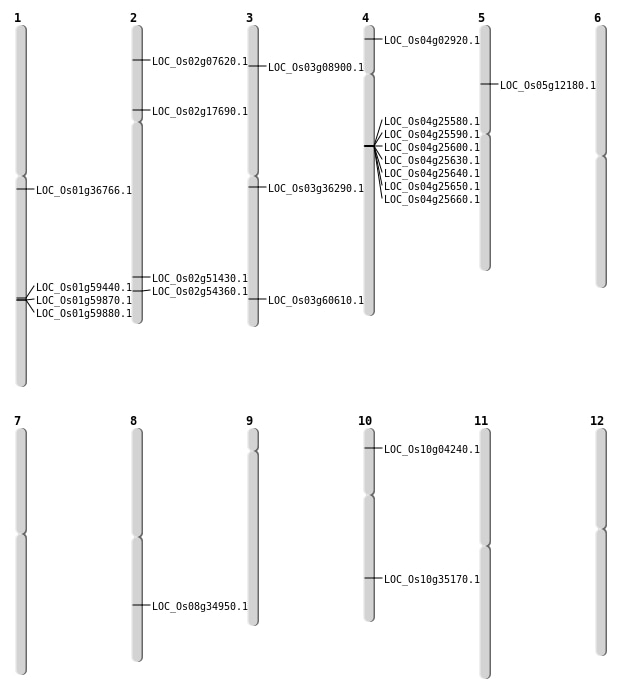
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17673622 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 20426340 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g36766 |
| SBS | Chr1 | 38105498 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 38105499 | G-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 762794 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1975894 | C-A | HOMO | STOP_GAINED | LOC_Os10g04240 |
| SBS | Chr12 | 19430946 | A-T | HOMO | UTR_5_PRIME | |
| SBS | Chr12 | 19430947 | A-T | HOMO | UTR_5_PRIME | |
| SBS | Chr12 | 6801941 | G-A | HET | INTRON | |
| SBS | Chr2 | 13887594 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 24728373 | T-A | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 3941277 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g07620 |
| SBS | Chr3 | 19056000 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 20128985 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g36290 |
| SBS | Chr3 | 20128987 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g36290 |
| SBS | Chr3 | 23478170 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 24117976 | G-A | HET | INTRON | |
| SBS | Chr4 | 1480339 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 16990451 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 18491080 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 21643270 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 25488994 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 6713288 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 6975857 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g12180 |
| SBS | Chr6 | 14105068 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 16196322 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 17198706 | A-C | HOMO | INTRON | |
| SBS | Chr6 | 22683825 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 26723607 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 11702965 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 11702966 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 12649161 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 14690660 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 21997814 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 28257549 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 14028313 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 14028314 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 26335308 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 14756658 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 17789824 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 7093626 | G-A | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1166028 | 1166033 | 5 | LOC_Os04g02920 |
| Deletion | Chr4 | 1344296 | 1344345 | 49 | |
| Deletion | Chr3 | 2159235 | 2159237 | 2 | |
| Deletion | Chr3 | 4614642 | 4614644 | 2 | LOC_Os03g08900 |
| Deletion | Chr1 | 5792267 | 5792270 | 3 | |
| Deletion | Chr3 | 6742725 | 6742732 | 7 | |
| Deletion | Chr12 | 8414368 | 8414369 | 1 | |
| Deletion | Chr11 | 9202795 | 9202804 | 9 | |
| Deletion | Chr2 | 9661316 | 9661318 | 2 | |
| Deletion | Chr3 | 9679632 | 9679657 | 25 | |
| Deletion | Chr1 | 12988920 | 12988925 | 5 | |
| Deletion | Chr4 | 14828001 | 14837000 | 9000 | 3 |
| Deletion | Chr4 | 14852001 | 14880000 | 28000 | 4 |
| Deletion | Chr3 | 17077639 | 17077651 | 12 | |
| Deletion | Chr4 | 17361372 | 17361373 | 1 | |
| Deletion | Chr1 | 17604430 | 17604440 | 10 | |
| Deletion | Chr9 | 17937882 | 17937887 | 5 | |
| Deletion | Chr10 | 18776566 | 18776574 | 8 | LOC_Os10g35170 |
| Deletion | Chr12 | 19483223 | 19483238 | 15 | |
| Deletion | Chr8 | 22003824 | 22003834 | 10 | LOC_Os08g34950 |
| Deletion | Chr6 | 27290150 | 27290151 | 1 | |
| Deletion | Chr2 | 31512386 | 31512392 | 6 | LOC_Os02g51430 |
| Deletion | Chr2 | 32361484 | 32361487 | 3 | |
| Deletion | Chr1 | 32811085 | 32811092 | 7 | |
| Deletion | Chr2 | 33321878 | 33321883 | 5 | LOC_Os02g54360 |
| Deletion | Chr1 | 34381370 | 34381384 | 14 | LOC_Os01g59440 |
| Deletion | Chr3 | 34440757 | 34440783 | 26 | LOC_Os03g60610 |
| Deletion | Chr1 | 34629632 | 34629640 | 8 | 2 |
Insertions: 7
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 10198415 | 10201376 | LOC_Os02g17690 |
No Translocation
