Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN836-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN836-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 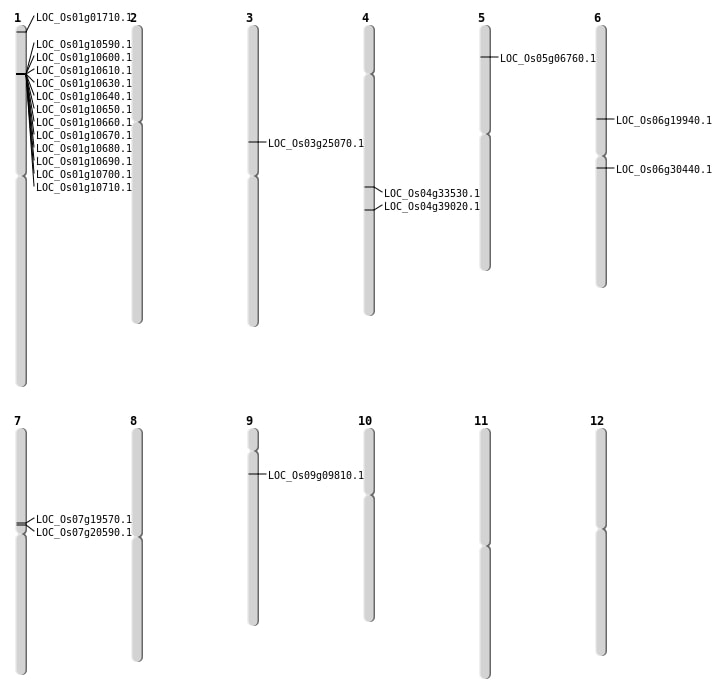
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24928219 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 28886204 | T-C | HET | INTRON | |
| SBS | Chr1 | 41529439 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 13505826 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 21998912 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 5208843 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 5720627 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 15167928 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 5848251 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 778173 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 14102713 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 14383284 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 26562552 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 15013716 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 34168419 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 151486 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 16088116 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 21449462 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 25521056 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 26663936 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 30215260 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 35710018 | T-C | HET | INTRON | |
| SBS | Chr4 | 12117622 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 19410072 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 20370104 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 8106489 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 12122345 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 18767926 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 24137532 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 2908679 | G-A | HET | INTRON | |
| SBS | Chr6 | 6959951 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 1123505 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 11598160 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g19570 |
| SBS | Chr7 | 14472131 | T-A | HOMO | UTR_5_PRIME | |
| SBS | Chr7 | 26287359 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 3651175 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 12915399 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 4035444 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 8265829 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 9191549 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 9968153 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 11483164 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 18006589 | T-C | HET | INTRON | |
| SBS | Chr9 | 5317800 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g09810 |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 377316 | 377317 | 1 | LOC_Os01g01710 |
| Deletion | Chr11 | 2397379 | 2397394 | 15 | |
| Deletion | Chr1 | 5650001 | 5736000 | 86000 | 12 |
| Deletion | Chr11 | 7264679 | 7264680 | 1 | |
| Deletion | Chr4 | 10450671 | 10450694 | 23 | |
| Deletion | Chr7 | 11569604 | 11569608 | 4 | |
| Deletion | Chr7 | 11903478 | 11903487 | 9 | LOC_Os07g20590 |
| Deletion | Chr11 | 14049829 | 14049848 | 19 | |
| Deletion | Chr10 | 14227566 | 14227573 | 7 | |
| Deletion | Chr12 | 16922297 | 16922302 | 5 | |
| Deletion | Chr6 | 17589720 | 17589721 | 1 | LOC_Os06g30440 |
| Deletion | Chr9 | 18166234 | 18166241 | 7 | |
| Deletion | Chr4 | 20933866 | 20933910 | 44 | |
| Deletion | Chr1 | 21041643 | 21041652 | 9 | |
| Deletion | Chr8 | 21721477 | 21721479 | 2 | |
| Deletion | Chr4 | 23175878 | 23175880 | 2 | LOC_Os04g39020 |
| Deletion | Chr4 | 25770805 | 25770811 | 6 | |
| Deletion | Chr6 | 27019146 | 27019147 | 1 | |
| Deletion | Chr5 | 27810379 | 27810393 | 14 | |
| Deletion | Chr7 | 29033481 | 29033482 | 1 | |
| Deletion | Chr5 | 29552125 | 29552126 | 1 | |
| Deletion | Chr1 | 34903021 | 34903028 | 7 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 15096802 | 15096802 | 1 | |
| Insertion | Chr11 | 23843690 | 23843690 | 1 | |
| Insertion | Chr3 | 14316094 | 14316095 | 2 | LOC_Os03g25070 |
| Insertion | Chr7 | 5268745 | 5268746 | 2 | |
| Insertion | Chr8 | 3347906 | 3347907 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 19964642 | 20284377 | LOC_Os04g33530 |
| Inversion | Chr4 | 19964653 | 20284375 | LOC_Os04g33530 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 11416701 | Chr5 | 3531246 | |
| Translocation | Chr6 | 11416791 | Chr5 | 3532826 | 2 |
