Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN849-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN849-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 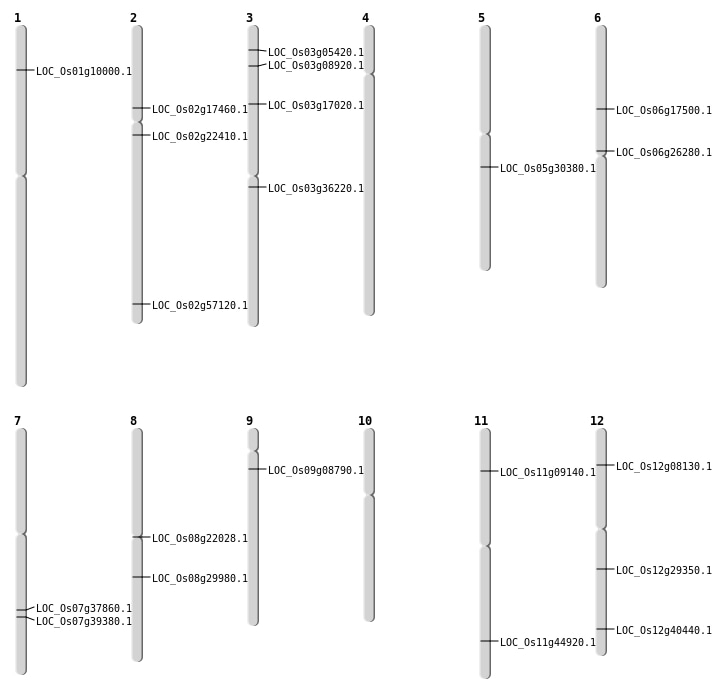
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12713104 | C-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 4395420 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 16301090 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 2097945 | A-T | HET | INTRON | |
| SBS | Chr11 | 27192193 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44920 |
| SBS | Chr11 | 6432397 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 13386087 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g22410 |
| SBS | Chr2 | 16030750 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 29401877 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 33577907 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 34985101 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g57120 |
| SBS | Chr3 | 10751825 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 14273179 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 20088799 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g36220 |
| SBS | Chr3 | 2679817 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g05420 |
| SBS | Chr3 | 32901516 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 4617233 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g08920 |
| SBS | Chr4 | 1528263 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 33424153 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 17608965 | G-A | HOMO | INTRON | |
| SBS | Chr5 | 26015142 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 6387073 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 20841954 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 22704614 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g37860 |
| SBS | Chr7 | 23590777 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g39380 |
| SBS | Chr7 | 9197305 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 13262546 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g22028 |
| SBS | Chr9 | 14633705 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 19310112 | C-T | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 437622 | 437635 | 13 | |
| Deletion | Chr9 | 627650 | 627651 | 1 | |
| Deletion | Chr4 | 2466343 | 2466344 | 1 | |
| Deletion | Chr7 | 3848497 | 3848509 | 12 | |
| Deletion | Chr12 | 4145961 | 4145962 | 1 | LOC_Os12g08130 |
| Deletion | Chr11 | 4868474 | 4868476 | 2 | LOC_Os11g09140 |
| Deletion | Chr1 | 5213195 | 5213203 | 8 | LOC_Os01g10000 |
| Deletion | Chr7 | 8554015 | 8554016 | 1 | |
| Deletion | Chr9 | 9433281 | 9433287 | 6 | |
| Deletion | Chr3 | 9455088 | 9455089 | 1 | LOC_Os03g17020 |
| Deletion | Chr7 | 9820080 | 9820085 | 5 | |
| Deletion | Chr1 | 14394822 | 14394841 | 19 | |
| Deletion | Chr7 | 14588553 | 14588577 | 24 | |
| Deletion | Chr6 | 15389296 | 15389302 | 6 | LOC_Os06g26280 |
| Deletion | Chr12 | 17416818 | 17416819 | 1 | LOC_Os12g29350 |
| Deletion | Chr5 | 17607312 | 17607327 | 15 | LOC_Os05g30380 |
| Deletion | Chr12 | 18366456 | 18366460 | 4 | |
| Deletion | Chr1 | 19108592 | 19108595 | 3 | |
| Deletion | Chr3 | 19664314 | 19664317 | 3 | |
| Deletion | Chr2 | 22444295 | 22444298 | 3 | |
| Deletion | Chr7 | 22703413 | 22703423 | 10 | |
| Deletion | Chr9 | 22777681 | 22777701 | 20 | |
| Deletion | Chr6 | 24480749 | 24480750 | 1 | |
| Deletion | Chr3 | 27192982 | 27192991 | 9 | |
| Deletion | Chr3 | 28889005 | 28889006 | 1 | |
| Deletion | Chr1 | 29244613 | 29244617 | 4 | |
| Deletion | Chr3 | 29653076 | 29653082 | 6 | |
| Deletion | Chr4 | 33999704 | 33999705 | 1 | |
| Deletion | Chr2 | 35847484 | 35847485 | 1 | |
| Deletion | Chr1 | 41506801 | 41506802 | 1 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 6051807 | 6051807 | 1 | |
| Insertion | Chr3 | 21100816 | 21100816 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 7747011 | 16310219 | |
| Inversion | Chr7 | 7747029 | 16310222 | |
| Inversion | Chr2 | 10042790 | 10501537 | LOC_Os02g17460 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 4416693 | Chr8 | 18415253 | LOC_Os08g29980 |
| Translocation | Chr9 | 4638141 | Chr5 | 14581433 | LOC_Os09g08790 |
| Translocation | Chr9 | 22821263 | Chr1 | 6563239 | |
| Translocation | Chr12 | 25027407 | Chr6 | 10152186 | 2 |
