Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN852-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN852-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 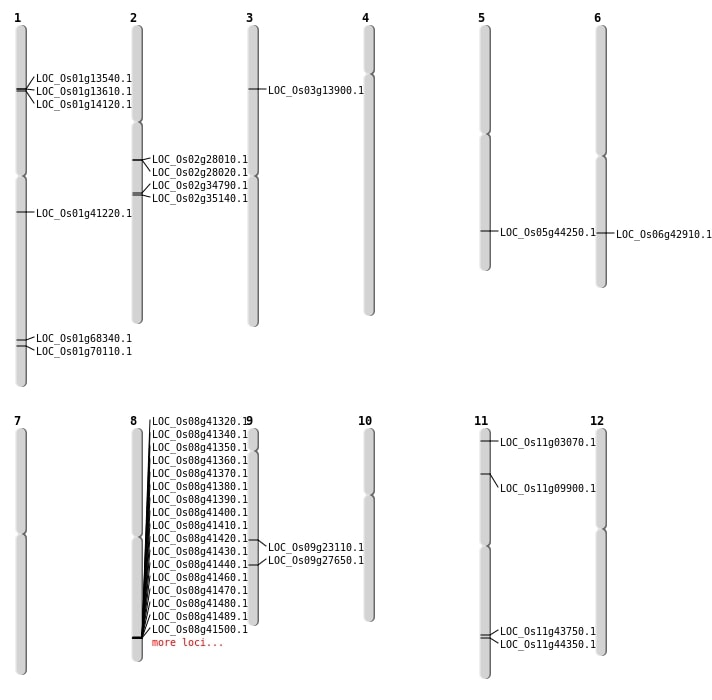
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12513187 | G-A | HET | INTRON | |
| SBS | Chr1 | 21428496 | C-G | HOMO | INTRON | |
| SBS | Chr1 | 35190609 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 39809283 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 6572858 | A-T | HET | INTRON | |
| SBS | Chr10 | 21862843 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 17613688 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 5299094 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g09900 |
| SBS | Chr12 | 24883047 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 27276729 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr12 | 27276732 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr12 | 4845945 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 7271488 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 21109547 | C-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 16773623 | G-A | HET | INTRON | |
| SBS | Chr3 | 22536250 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 34016613 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 7537612 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g13900 |
| SBS | Chr4 | 18391879 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 21046542 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 23414184 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 4655704 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 18899674 | G-A | HET | INTRON | |
| SBS | Chr5 | 21140974 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 24444068 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr5 | 5444859 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 7797539 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 16123567 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 27504762 | C-T | HET | INTRON | |
| SBS | Chr7 | 28201663 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 11290576 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 13769139 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 16174340 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 11334504 | C-T | HET | SYNONYMOUS_CODING |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 147290 | 147314 | 24 | |
| Deletion | Chr11 | 1087318 | 1087328 | 10 | LOC_Os11g03070 |
| Deletion | Chr8 | 2374971 | 2374985 | 14 | |
| Deletion | Chr11 | 2877141 | 2877143 | 2 | |
| Deletion | Chr5 | 2923989 | 2924000 | 11 | |
| Deletion | Chr6 | 5574035 | 5574047 | 12 | |
| Deletion | Chr5 | 6254401 | 6254407 | 6 | |
| Deletion | Chr1 | 7924753 | 7924764 | 11 | LOC_Os01g14120 |
| Deletion | Chr10 | 11614900 | 11614901 | 1 | |
| Deletion | Chr2 | 11644663 | 11644664 | 1 | |
| Deletion | Chr9 | 13666899 | 13666900 | 1 | LOC_Os09g23110 |
| Deletion | Chr12 | 15527284 | 15527285 | 1 | |
| Deletion | Chr7 | 15646355 | 15646356 | 1 | |
| Deletion | Chr2 | 16582001 | 16592000 | 10000 | 2 |
| Deletion | Chr9 | 16825431 | 16825432 | 1 | LOC_Os09g27650 |
| Deletion | Chr9 | 20466592 | 20466597 | 5 | |
| Deletion | Chr1 | 20957092 | 20957109 | 17 | |
| Deletion | Chr8 | 21935703 | 21935706 | 3 | |
| Deletion | Chr1 | 23337028 | 23337029 | 1 | LOC_Os01g41220 |
| Deletion | Chr5 | 25742118 | 25747011 | 4893 | LOC_Os05g44250 |
| Deletion | Chr6 | 25786630 | 25786632 | 2 | LOC_Os06g42910 |
| Deletion | Chr8 | 26098199 | 26426595 | 328396 | 50 |
| Deletion | Chr3 | 34987532 | 34987937 | 405 | |
| Deletion | Chr1 | 35127346 | 35127350 | 4 | |
| Deletion | Chr1 | 39197105 | 39197112 | 7 | |
| Deletion | Chr1 | 39709855 | 39710924 | 1069 | LOC_Os01g68340 |
| Deletion | Chr1 | 40572961 | 40572963 | 2 | LOC_Os01g70110 |
Insertions: 6
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 7572652 | 7621122 | 2 |
| Inversion | Chr1 | 7577335 | 7621122 | LOC_Os01g13610 |
| Inversion | Chr5 | 13959699 | 20573839 | |
| Inversion | Chr1 | 19971917 | 35935329 | |
| Inversion | Chr1 | 19979703 | 35935316 | |
| Inversion | Chr2 | 20858199 | 21102619 | 2 |
| Inversion | Chr11 | 26420799 | 26810428 | 2 |
| Inversion | Chr11 | 26810441 | 26938172 | LOC_Os11g44350 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 11452205 | Chr1 | 39710093 | |
| Translocation | Chr12 | 13878878 | Chr11 | 26424908 | |
| Translocation | Chr12 | 13878884 | Chr11 | 26420813 | LOC_Os11g43750 |
