Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN863-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN863-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 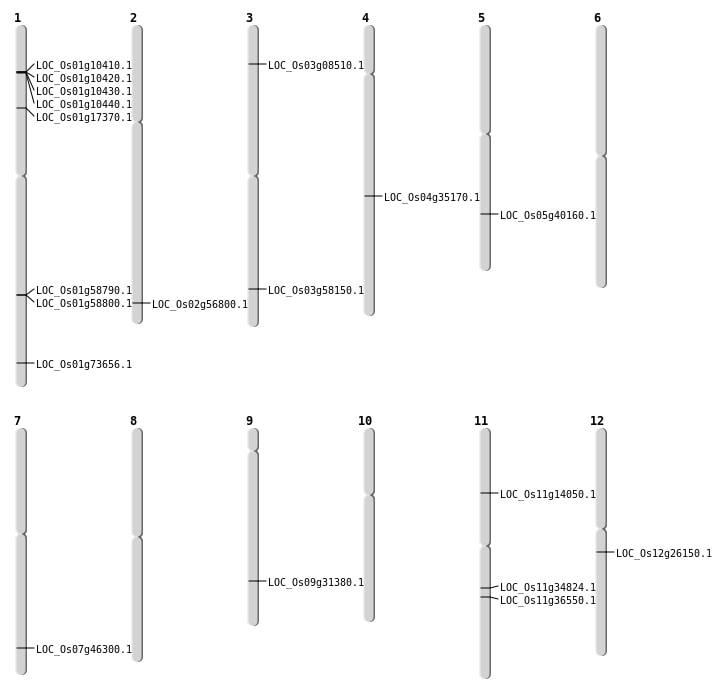
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12434342 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 26363480 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 29825753 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 36930650 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 9999549 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g17370 |
| SBS | Chr11 | 20404645 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g34824 |
| SBS | Chr11 | 21561929 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 8521128 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 15235441 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26150 |
| SBS | Chr2 | 18530418 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 18738977 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 18861906 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr2 | 34814220 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g56800 |
| SBS | Chr2 | 34918073 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 16340082 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 31501792 | G-A | HET | INTRON | |
| SBS | Chr3 | 34105950 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 7568065 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 7746068 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 10268270 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 23610392 | A-G | HET | INTRON | |
| SBS | Chr5 | 9588499 | C-A | HET | INTRON | |
| SBS | Chr6 | 1481438 | C-T | HET | INTRON | |
| SBS | Chr6 | 2496126 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 25571408 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 2617627 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 28933990 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 18008446 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 27623128 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g46300 |
| SBS | Chr7 | 8255683 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 8255684 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 8834459 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 11413928 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 12027253 | C-T | HET | INTRON | |
| SBS | Chr9 | 12930967 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 14249636 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 18867078 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g31380 |
| SBS | Chr9 | 802053 | A-T | HET | INTRON |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1689677 | 1689678 | 1 | |
| Deletion | Chr11 | 2067057 | 2067078 | 21 | |
| Deletion | Chr3 | 4377963 | 4377973 | 10 | LOC_Os03g08510 |
| Deletion | Chr5 | 5348488 | 5348491 | 3 | |
| Deletion | Chr1 | 5480001 | 5508000 | 28000 | 4 |
| Deletion | Chr11 | 7798054 | 7798070 | 16 | LOC_Os11g14050 |
| Deletion | Chr2 | 9028158 | 9028161 | 3 | |
| Deletion | Chr12 | 10289809 | 10289814 | 5 | |
| Deletion | Chr5 | 10418867 | 10418876 | 9 | |
| Deletion | Chr9 | 11777545 | 11777551 | 6 | |
| Deletion | Chr7 | 12775851 | 12775852 | 1 | |
| Deletion | Chr11 | 13388733 | 13388736 | 3 | |
| Deletion | Chr7 | 14484661 | 14484670 | 9 | |
| Deletion | Chr2 | 17236813 | 17236816 | 3 | |
| Deletion | Chr5 | 18189315 | 18189328 | 13 | |
| Deletion | Chr3 | 20216383 | 20216384 | 1 | |
| Deletion | Chr5 | 21244165 | 21244172 | 7 | |
| Deletion | Chr11 | 21561987 | 21562007 | 20 | LOC_Os11g36550 |
| Deletion | Chr6 | 23828703 | 23828704 | 1 | |
| Deletion | Chr8 | 24457867 | 24457868 | 1 | |
| Deletion | Chr2 | 25030021 | 25030023 | 2 | |
| Deletion | Chr6 | 25889473 | 25889475 | 2 | |
| Deletion | Chr12 | 27153233 | 27153238 | 5 | |
| Deletion | Chr4 | 30152645 | 30152653 | 8 | |
| Deletion | Chr3 | 33103263 | 33103265 | 2 | LOC_Os03g58150 |
| Deletion | Chr1 | 33976660 | 33976661 | 1 | 2 |
| Deletion | Chr3 | 34112712 | 34112715 | 3 | |
| Deletion | Chr3 | 35263144 | 35263168 | 24 | |
| Deletion | Chr1 | 42677937 | 42677939 | 2 | LOC_Os01g73656 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 11342124 | 11342124 | 1 | |
| Insertion | Chr4 | 21374534 | 21374534 | 1 | LOC_Os04g35170 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 8928660 | 15935317 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 27797888 | Chr5 | 23590043 | LOC_Os05g40160 |
