Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN887-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN887-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 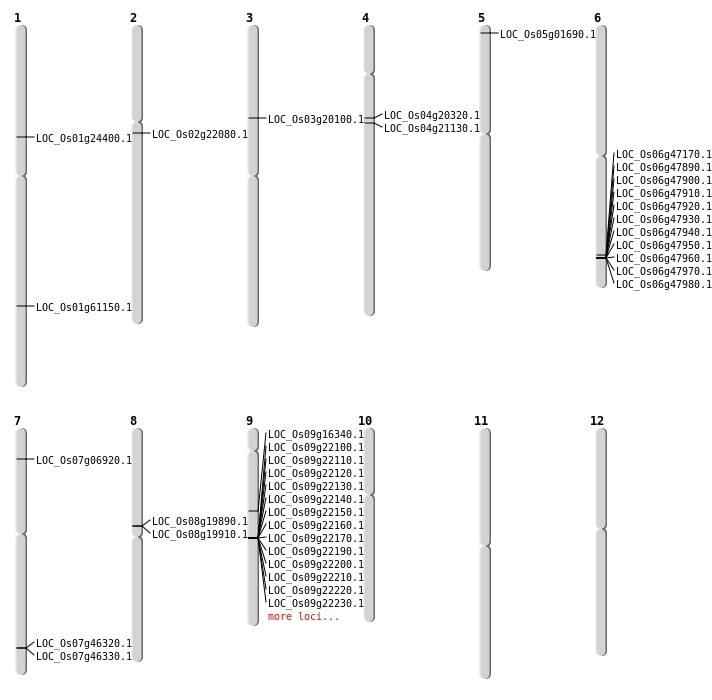
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13754306 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g24400 |
| SBS | Chr1 | 22883669 | A-G | HET | INTRON | |
| SBS | Chr1 | 34661457 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 10614113 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 20026253 | A-G | HET | INTRON | |
| SBS | Chr10 | 20129394 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 20306528 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 9130924 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 2373717 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 2373718 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 9495132 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 25078074 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 840649 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 246277 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 26794939 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 12622492 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 34352986 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 14317018 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 4005652 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 419412 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr6 | 12672722 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 17652576 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 28604747 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g47170 |
| SBS | Chr7 | 27443068 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 29424073 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 3383770 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g06920 |
| SBS | Chr7 | 3998447 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 8329575 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 8324541 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 13805119 | C-G | HOMO | INTRON | |
| SBS | Chr9 | 1518499 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 8231003 | G-A | HOMO | SYNONYMOUS_CODING |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 286108 | 286114 | 6 | |
| Deletion | Chr5 | 419246 | 419250 | 4 | LOC_Os05g01690 |
| Deletion | Chr11 | 2008284 | 2008294 | 10 | |
| Deletion | Chr5 | 2547934 | 2547949 | 15 | |
| Deletion | Chr7 | 3373082 | 3373105 | 23 | |
| Deletion | Chr10 | 3831059 | 3831060 | 1 | |
| Deletion | Chr8 | 6004292 | 6004293 | 1 | |
| Deletion | Chr10 | 7119307 | 7119308 | 1 | |
| Deletion | Chr1 | 8446689 | 8446692 | 3 | |
| Deletion | Chr11 | 9073021 | 9073027 | 6 | |
| Deletion | Chr9 | 9990843 | 9990846 | 3 | LOC_Os09g16340 |
| Deletion | Chr6 | 10418749 | 10418756 | 7 | |
| Deletion | Chr4 | 11897960 | 11897972 | 12 | LOC_Os04g21130 |
| Deletion | Chr8 | 11911618 | 11911651 | 33 | LOC_Os08g19890 |
| Deletion | Chr8 | 11927075 | 11927076 | 1 | LOC_Os08g19910 |
| Deletion | Chr2 | 13133790 | 13133791 | 1 | LOC_Os02g22080 |
| Deletion | Chr9 | 13389001 | 13704000 | 315000 | 53 |
| Deletion | Chr11 | 14676954 | 14676975 | 21 | |
| Deletion | Chr12 | 17807641 | 17807655 | 14 | |
| Deletion | Chr7 | 19035604 | 19035618 | 14 | |
| Deletion | Chr7 | 19415308 | 19415311 | 3 | |
| Deletion | Chr6 | 23066319 | 23066327 | 8 | |
| Deletion | Chr6 | 27763764 | 27763765 | 1 | |
| Deletion | Chr6 | 28974001 | 29012000 | 38000 | 10 |
| Deletion | Chr3 | 31301182 | 31301185 | 3 | |
| Deletion | Chr2 | 34048539 | 34048542 | 3 | |
| Deletion | Chr1 | 35396297 | 35396298 | 1 | LOC_Os01g61150 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 18804838 | 18804841 | 4 | |
| Insertion | Chr4 | 20701596 | 20701597 | 2 | |
| Insertion | Chr5 | 6265174 | 6265175 | 2 | |
| Insertion | Chr6 | 5232124 | 5232124 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 27628527 | 27631421 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 4638822 | Chr2 | 26867153 | LOC_Os02g44380 |
| Translocation | Chr10 | 10201519 | Chr2 | 26867154 | LOC_Os02g44380 |
| Translocation | Chr3 | 11353288 | Chr2 | 4182465 | LOC_Os03g20100 |
| Translocation | Chr4 | 11367242 | Chr1 | 33407001 | LOC_Os04g20320 |
| Translocation | Chr4 | 11367567 | Chr1 | 33407001 | LOC_Os04g20320 |
| Translocation | Chr9 | 13394044 | Chr2 | 4182465 | 2 |
| Translocation | Chr9 | 13397008 | Chr2 | 4182450 | LOC_Os09g22120 |
