Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN891-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN891-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 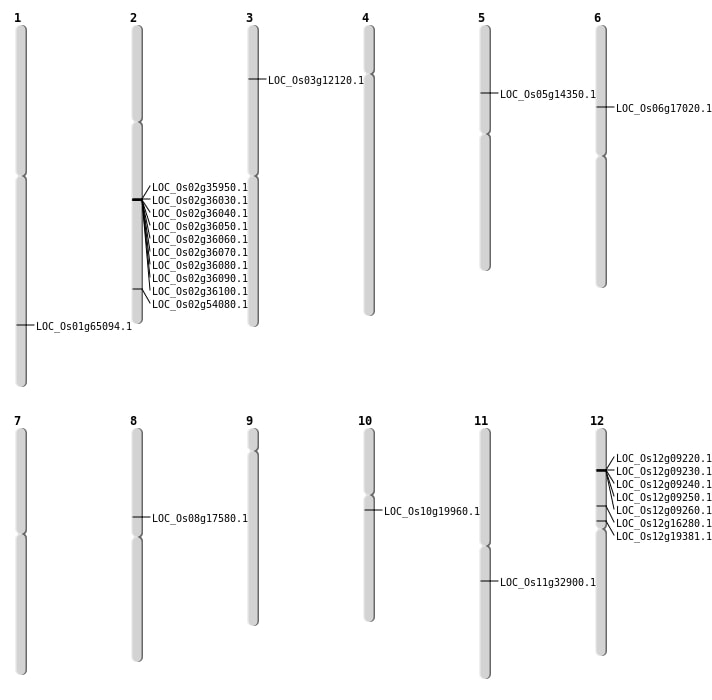
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 27654293 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 16580956 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 11106939 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 15045217 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 19039340 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 24896147 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 634190 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 16557844 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 25761039 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 4858583 | T-G | HET | INTRON | |
| SBS | Chr2 | 12790139 | A-G | HET | INTRON | |
| SBS | Chr2 | 13612169 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 16889705 | C-T | HET | INTRON | |
| SBS | Chr2 | 20282309 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 20797760 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 4255288 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 4802991 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr4 | 7257868 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 8196296 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 15786623 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 18575184 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 3739891 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 3739892 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 8074271 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g14350 |
| SBS | Chr6 | 16386596 | G-T | HET | INTRON | |
| SBS | Chr6 | 24748850 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 5320001 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 5320002 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 9709265 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 10616253 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 25485911 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2625305 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 28846562 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 410532 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 20054308 | A-G | HOMO | INTRON | |
| SBS | Chr9 | 22456023 | T-A | HOMO | INTRON | |
| SBS | Chr9 | 3548093 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 4979645 | G-C | HET | UTR_5_PRIME |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 74490 | 74491 | 1 | |
| Deletion | Chr3 | 1115880 | 1115895 | 15 | |
| Deletion | Chr4 | 1731563 | 1731576 | 13 | |
| Deletion | Chr3 | 4664825 | 4664827 | 2 | |
| Deletion | Chr8 | 4778428 | 4778429 | 1 | |
| Deletion | Chr12 | 4812001 | 4856000 | 44000 | 5 |
| Deletion | Chr1 | 5093444 | 5093453 | 9 | |
| Deletion | Chr3 | 6360005 | 6360010 | 5 | LOC_Os03g12120 |
| Deletion | Chr12 | 9301883 | 9301885 | 2 | LOC_Os12g16280 |
| Deletion | Chr6 | 9872316 | 9872317 | 1 | LOC_Os06g17020 |
| Deletion | Chr10 | 10014175 | 10014186 | 11 | LOC_Os10g19960 |
| Deletion | Chr7 | 10615641 | 10615670 | 29 | |
| Deletion | Chr8 | 11913528 | 11913529 | 1 | |
| Deletion | Chr10 | 13622627 | 13622628 | 1 | |
| Deletion | Chr4 | 16063992 | 16063994 | 2 | |
| Deletion | Chr9 | 17032601 | 17032611 | 10 | |
| Deletion | Chr7 | 17220059 | 17220060 | 1 | |
| Deletion | Chr11 | 19323129 | 19323135 | 6 | |
| Deletion | Chr8 | 19620989 | 19620998 | 9 | |
| Deletion | Chr2 | 20810345 | 20810346 | 1 | |
| Deletion | Chr12 | 21247509 | 21247511 | 2 | |
| Deletion | Chr2 | 21646001 | 21727000 | 81000 | 8 |
| Deletion | Chr9 | 22446886 | 22446887 | 1 | |
| Deletion | Chr1 | 24167220 | 24167221 | 1 | |
| Deletion | Chr6 | 25403053 | 25403103 | 50 | |
| Deletion | Chr4 | 31164146 | 31164148 | 2 | |
| Deletion | Chr1 | 32973128 | 32973133 | 5 | |
| Deletion | Chr1 | 37785394 | 37785396 | 2 | LOC_Os01g65094 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 20665523 | 20665523 | 1 | |
| Insertion | Chr2 | 17969345 | 17969346 | 2 | |
| Insertion | Chr5 | 16375533 | 16375534 | 2 | |
| Insertion | Chr5 | 4844316 | 4844317 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 15096305 | 18138985 | |
| Inversion | Chr2 | 21601738 | 21727805 | 2 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 10754470 | Chr6 | 19260595 | LOC_Os08g17580 |
| Translocation | Chr12 | 11272892 | Chr9 | 21454316 | LOC_Os12g19381 |
| Translocation | Chr11 | 19434893 | Chr2 | 33125349 | 2 |
