Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN897-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN897-S Alignment File |
| Seed Availability | No |
| Mapping (Hover to Zoom-In) | 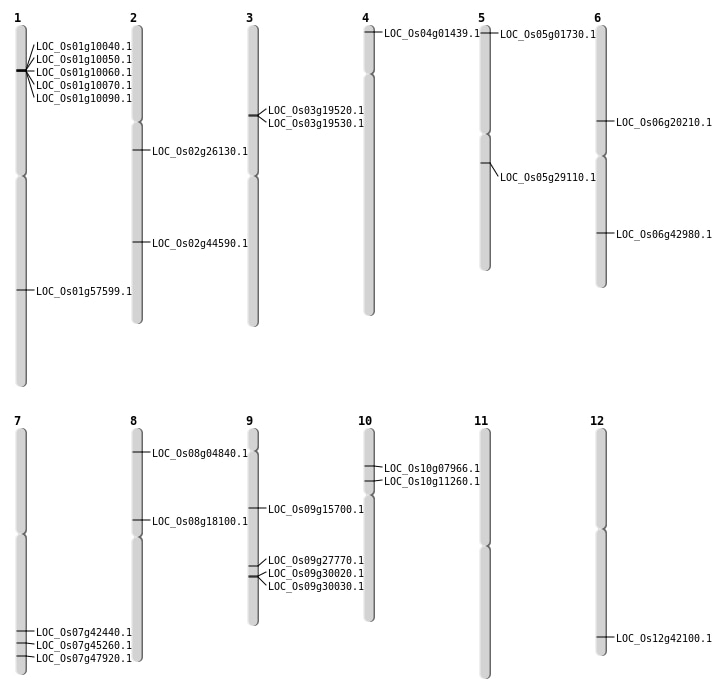
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21655012 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 13397231 | C-A | HET | INTRON | |
| SBS | Chr10 | 3866302 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 4245072 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g07966 |
| SBS | Chr10 | 4539377 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 21454551 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 22147999 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 22347169 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 27589543 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 7466577 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 10988430 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 15076670 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 16145161 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 21196461 | T-A | HET | INTRON | |
| SBS | Chr3 | 21550227 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 31590530 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 33454043 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 34066233 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 27331286 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 29279097 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 4431993 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 5912 | G-A | HET | INTRON | |
| SBS | Chr4 | 7938684 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 22344725 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 7879630 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 11598457 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g20210 |
| SBS | Chr6 | 23800533 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 25822391 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g42980 |
| SBS | Chr7 | 26110332 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 27228748 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 8588492 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 11098165 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g18100 |
| SBS | Chr8 | 17604534 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 1867446 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 20389512 | T-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 17071614 | T-C | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1752506 | 1752514 | 8 | |
| Deletion | Chr8 | 1785977 | 1785978 | 1 | |
| Deletion | Chr8 | 2434013 | 2434016 | 3 | LOC_Os08g04840 |
| Deletion | Chr8 | 3399714 | 3399727 | 13 | |
| Deletion | Chr10 | 3793461 | 3793475 | 14 | |
| Deletion | Chr1 | 5242914 | 5262289 | 19375 | 6 |
| Deletion | Chr2 | 6224195 | 6224202 | 7 | |
| Deletion | Chr10 | 6250799 | 6250824 | 25 | LOC_Os10g11260 |
| Deletion | Chr9 | 9926045 | 9926053 | 8 | |
| Deletion | Chr4 | 12383881 | 12383908 | 27 | |
| Deletion | Chr10 | 13296289 | 13296292 | 3 | |
| Deletion | Chr4 | 14209298 | 14209303 | 5 | |
| Deletion | Chr2 | 14352500 | 14352506 | 6 | |
| Deletion | Chr4 | 16519435 | 16519440 | 5 | |
| Deletion | Chr9 | 16897142 | 16897143 | 1 | LOC_Os09g27770 |
| Deletion | Chr9 | 18259463 | 18269170 | 9707 | 2 |
| Deletion | Chr8 | 19276346 | 19276347 | 1 | |
| Deletion | Chr3 | 20033468 | 20033475 | 7 | |
| Deletion | Chr1 | 21409211 | 21409217 | 6 | |
| Deletion | Chr12 | 21872597 | 21872600 | 3 | |
| Deletion | Chr1 | 23854567 | 23854570 | 3 | |
| Deletion | Chr12 | 26106422 | 26106423 | 1 | LOC_Os12g42100 |
| Deletion | Chr2 | 27020721 | 27020726 | 5 | LOC_Os02g44590 |
| Deletion | Chr1 | 27134421 | 27134428 | 7 | |
| Deletion | Chr6 | 27215895 | 27215899 | 4 | |
| Deletion | Chr7 | 28620342 | 28620550 | 208 | LOC_Os07g47920 |
| Deletion | Chr3 | 32309854 | 32309855 | 1 | |
| Deletion | Chr3 | 33427080 | 33427091 | 11 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11739696 | 11739699 | 4 | |
| Insertion | Chr1 | 33303789 | 33303789 | 1 | LOC_Os01g57599 |
| Insertion | Chr12 | 27134457 | 27134460 | 4 | |
| Insertion | Chr3 | 32575446 | 32575446 | 1 | |
| Insertion | Chr3 | 32950453 | 32950454 | 2 | |
| Insertion | Chr5 | 21765747 | 21765750 | 4 | |
| Insertion | Chr5 | 29627689 | 29627692 | 4 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 308754 | 1087425 | LOC_Os04g01439 |
| Inversion | Chr2 | 3032854 | 17852685 | |
| Inversion | Chr9 | 9434550 | 9578681 | LOC_Os09g15700 |
| Inversion | Chr9 | 9578686 | 14526956 | LOC_Os09g15700 |
| Inversion | Chr5 | 15866057 | 17096989 | LOC_Os05g29110 |
| Inversion | Chr5 | 16183344 | 17096921 | LOC_Os05g29110 |
| Inversion | Chr11 | 28440204 | 28440390 |
Translocations: 14
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 449000 | Chr3 | 10988075 | 2 |
| Translocation | Chr5 | 449001 | Chr3 | 10977806 | 2 |
| Translocation | Chr12 | 1286706 | Chr2 | 15353558 | LOC_Os02g26130 |
| Translocation | Chr10 | 2161990 | Chr1 | 5243200 | LOC_Os01g10040 |
| Translocation | Chr10 | 2166305 | Chr1 | 5262353 | LOC_Os01g10090 |
| Translocation | Chr5 | 15865971 | Chr1 | 5242913 | LOC_Os01g10040 |
| Translocation | Chr5 | 17096917 | Chr1 | 5243196 | 2 |
| Translocation | Chr9 | 18268281 | Chr7 | 27006693 | LOC_Os07g45260 |
| Translocation | Chr9 | 18269155 | Chr7 | 27006692 | 2 |
| Translocation | Chr3 | 22942522 | Chr1 | 39679149 | |
| Translocation | Chr11 | 27827885 | Chr7 | 25412078 | LOC_Os07g42440 |
| Translocation | Chr7 | 28620355 | Chr3 | 3374800 | LOC_Os07g47920 |
| Translocation | Chr7 | 28620541 | Chr3 | 3374798 | LOC_Os07g47920 |
| Translocation | Chr3 | 34354293 | Chr2 | 18055340 |
