Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN901-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN901-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 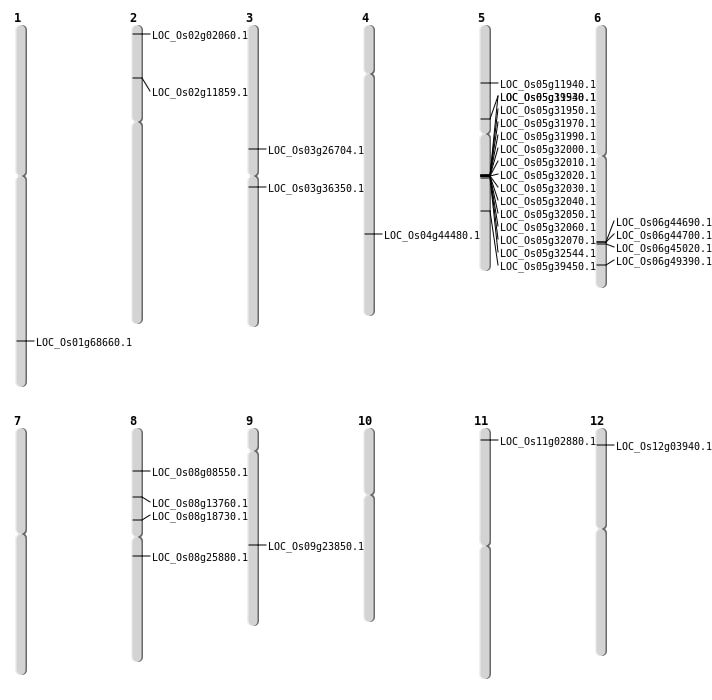
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12474874 | T-C | HET | INTRON | |
| SBS | Chr1 | 28962461 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 31032108 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 40538178 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 8792102 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr10 | 1699819 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 9590013 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 16153168 | G-A | HET | INTRON | |
| SBS | Chr11 | 976166 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g02880 |
| SBS | Chr12 | 19758060 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 20403547 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 13420603 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 18699629 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 28984926 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 31789385 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 14547660 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 15142001 | T-A | HET | INTRON | |
| SBS | Chr4 | 28698906 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 32705677 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 2117431 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 22022685 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 28317162 | A-C | HET | INTRON | |
| SBS | Chr5 | 3579783 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 5420611 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr5 | 8362162 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 9894496 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 21793424 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 26842213 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 26959456 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 16789412 | C-T | HET | INTRON | |
| SBS | Chr7 | 9124012 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 2737393 | T-G | HOMO | INTRON | |
| SBS | Chr8 | 8203753 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g13760 |
| SBS | Chr9 | 11084551 | C-G | HET | INTRON | |
| SBS | Chr9 | 12057989 | C-T | HET | INTRON | |
| SBS | Chr9 | 14222759 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g23850 |
| SBS | Chr9 | 17170804 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 22138051 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 4296779 | T-C | HET | INTRON |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 161943 | 161946 | 3 | |
| Deletion | Chr10 | 279240 | 279244 | 4 | |
| Deletion | Chr10 | 1085098 | 1085108 | 10 | |
| Deletion | Chr12 | 1290015 | 1290023 | 8 | |
| Deletion | Chr10 | 1795812 | 1795813 | 1 | |
| Deletion | Chr10 | 3398709 | 3398725 | 16 | |
| Deletion | Chr8 | 4934703 | 4934717 | 14 | LOC_Os08g08550 |
| Deletion | Chr2 | 6146143 | 6146144 | 1 | LOC_Os02g11859 |
| Deletion | Chr5 | 6794963 | 6794970 | 7 | LOC_Os05g11940 |
| Deletion | Chr3 | 6838868 | 6838869 | 1 | |
| Deletion | Chr8 | 7504235 | 7504236 | 1 | |
| Deletion | Chr4 | 8512337 | 8512339 | 2 | |
| Deletion | Chr3 | 8656005 | 8656015 | 10 | |
| Deletion | Chr2 | 8799116 | 8799117 | 1 | |
| Deletion | Chr8 | 11183956 | 11183971 | 15 | LOC_Os08g18730 |
| Deletion | Chr4 | 15312921 | 15312934 | 13 | |
| Deletion | Chr3 | 15595948 | 15595950 | 2 | |
| Deletion | Chr5 | 18588676 | 18588677 | 1 | |
| Deletion | Chr5 | 18606001 | 18697000 | 91000 | 12 |
| Deletion | Chr3 | 20008444 | 20008452 | 8 | |
| Deletion | Chr2 | 20120951 | 20120961 | 10 | |
| Deletion | Chr3 | 20157168 | 20157173 | 5 | LOC_Os03g36350 |
| Deletion | Chr1 | 20990948 | 20990950 | 2 | |
| Deletion | Chr7 | 21173916 | 21173922 | 6 | |
| Deletion | Chr5 | 23139216 | 23139230 | 14 | LOC_Os05g39450 |
| Deletion | Chr1 | 26307667 | 26307669 | 2 | |
| Deletion | Chr4 | 26326724 | 26326729 | 5 | LOC_Os04g44480 |
| Deletion | Chr6 | 26978001 | 26987000 | 9000 | 2 |
| Deletion | Chr2 | 27223355 | 27223362 | 7 | |
| Deletion | Chr6 | 27493991 | 27494001 | 10 | |
| Deletion | Chr8 | 27625588 | 27625590 | 2 | |
| Deletion | Chr6 | 29927144 | 29927157 | 13 | LOC_Os06g49390 |
| Deletion | Chr2 | 30866048 | 30866053 | 5 | |
| Deletion | Chr1 | 39876848 | 39876855 | 7 | LOC_Os01g68660 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 20273415 | 20273416 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 19022648 | 20756129 | LOC_Os05g32544 |
| Inversion | Chr6 | 26986758 | 27228213 | LOC_Os06g45020 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 1631234 | Chr5 | 7511900 | LOC_Os12g03940 |
| Translocation | Chr12 | 3661296 | Chr3 | 33837597 | |
| Translocation | Chr12 | 3662760 | Chr3 | 33837520 | |
| Translocation | Chr12 | 3662764 | Chr5 | 18696837 | |
| Translocation | Chr12 | 3668689 | Chr6 | 27228216 | LOC_Os06g45020 |
| Translocation | Chr12 | 9066726 | Chr9 | 12080317 | |
| Translocation | Chr5 | 11351935 | Chr2 | 604382 | 2 |
| Translocation | Chr8 | 15765017 | Chr3 | 15248515 | 2 |
| Translocation | Chr4 | 31353352 | Chr2 | 32187635 |
