Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN902-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN902-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 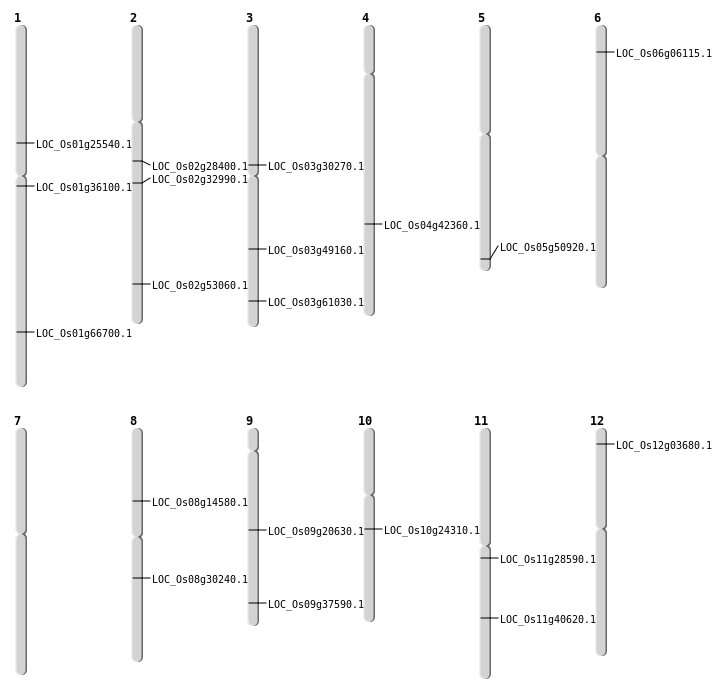
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25084005 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 27040217 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 22038203 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16485837 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g28590 |
| SBS | Chr12 | 4621959 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 3974970 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 4760785 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 19812115 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 28003447 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g49160 |
| SBS | Chr4 | 16036414 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 16117632 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 25064519 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g42360 |
| SBS | Chr4 | 25731148 | T-G | HET | INTRON | |
| SBS | Chr4 | 26126415 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 30934281 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 33635367 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 478002 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 29223059 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g50920 |
| SBS | Chr5 | 3902137 | C-T | HOMO | INTRON | |
| SBS | Chr6 | 9352081 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 29075578 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr7 | 4169436 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 11590828 | T-A | HOMO | UTR_3_PRIME | |
| SBS | Chr8 | 11590834 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr8 | 17271856 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 1846371 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 25431973 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 11997854 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 21673464 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g37590 |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 5654673 | 5654695 | 22 | |
| Deletion | Chr11 | 7629947 | 7629948 | 1 | |
| Deletion | Chr8 | 8768378 | 8768380 | 2 | LOC_Os08g14580 |
| Deletion | Chr2 | 10133350 | 10133369 | 19 | |
| Deletion | Chr8 | 11596008 | 11596021 | 13 | |
| Deletion | Chr11 | 11792489 | 11792505 | 16 | |
| Deletion | Chr1 | 14480894 | 14480903 | 9 | LOC_Os01g25540 |
| Deletion | Chr2 | 19614721 | 19614728 | 7 | LOC_Os02g32990 |
| Deletion | Chr6 | 21006527 | 21006548 | 21 | |
| Deletion | Chr10 | 22038205 | 22038209 | 4 | |
| Deletion | Chr11 | 24247604 | 24247634 | 30 | LOC_Os11g40620 |
| Deletion | Chr12 | 25575982 | 25575996 | 14 | |
| Deletion | Chr6 | 28743255 | 28743256 | 1 | |
| Deletion | Chr3 | 34351044 | 34351045 | 1 | |
| Deletion | Chr3 | 34674127 | 34674128 | 1 | LOC_Os03g61030 |
No Insertion
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 12420348 | 12429000 | LOC_Os09g20630 |
| Inversion | Chr9 | 12420359 | 12429009 | LOC_Os09g20630 |
| Inversion | Chr2 | 32466522 | 32753198 | LOC_Os02g53060 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 754123 | Chr9 | 20686660 | |
| Translocation | Chr12 | 1482618 | Chr2 | 9635649 | LOC_Os12g03680 |
| Translocation | Chr5 | 4616088 | Chr3 | 5945889 | |
| Translocation | Chr10 | 12452273 | Chr3 | 13548341 | LOC_Os10g24310 |
| Translocation | Chr3 | 17277958 | Chr2 | 16784615 | 2 |
| Translocation | Chr8 | 18582574 | Chr1 | 19977624 | 2 |
| Translocation | Chr12 | 19376806 | Chr1 | 14643382 | |
| Translocation | Chr8 | 27374158 | Chr6 | 2829720 | LOC_Os06g06115 |
| Translocation | Chr2 | 32751794 | Chr1 | 38740725 | LOC_Os01g66700 |
| Translocation | Chr2 | 32753003 | Chr1 | 38740721 | LOC_Os01g66700 |
