Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN903-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN903-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 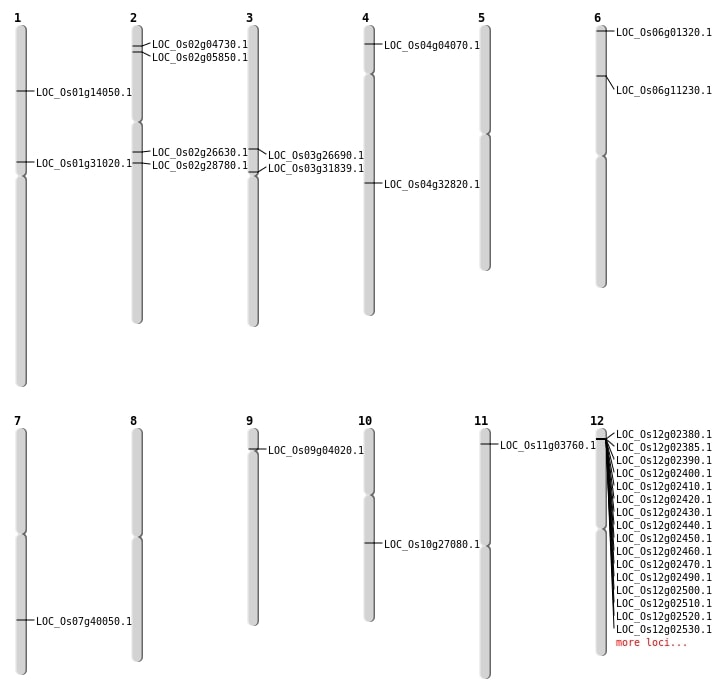
|
| Variant Information |
|---|
Single base substitutions: 22
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24459524 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 7863400 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g14050 |
| SBS | Chr10 | 20285965 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 3078053 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 24831538 | C-T | HET | INTRON | |
| SBS | Chr3 | 19346358 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 24361482 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 26434709 | T-A | HET | INTRON | |
| SBS | Chr4 | 19565330 | A-T | HET | INTRON | |
| SBS | Chr4 | 32478404 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 10148581 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 17214284 | T-A | HET | INTRON | |
| SBS | Chr5 | 3917381 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 568387 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 7233738 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 25707239 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 26469474 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 30403502 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 13692108 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 22781323 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 5214588 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 2792324 | A-C | HET | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 3381 | 3382 | 1 | |
| Deletion | Chr6 | 204261 | 204264 | 3 | LOC_Os06g01320 |
| Deletion | Chr12 | 779001 | 907000 | 128000 | 24 |
| Deletion | Chr2 | 1463407 | 1463411 | 4 | |
| Deletion | Chr4 | 1879442 | 1879457 | 15 | LOC_Os04g04070 |
| Deletion | Chr2 | 2889276 | 2889280 | 4 | LOC_Os02g05850 |
| Deletion | Chr1 | 3529749 | 3529757 | 8 | |
| Deletion | Chr12 | 4222777 | 4222778 | 1 | |
| Deletion | Chr12 | 7348647 | 7348652 | 5 | |
| Deletion | Chr12 | 8349767 | 8349768 | 1 | |
| Deletion | Chr2 | 9109234 | 9109252 | 18 | |
| Deletion | Chr1 | 9300345 | 9300353 | 8 | |
| Deletion | Chr7 | 14303328 | 14303329 | 1 | |
| Deletion | Chr7 | 15393785 | 15393787 | 2 | |
| Deletion | Chr1 | 16966967 | 16966975 | 8 | LOC_Os01g31020 |
| Deletion | Chr5 | 20910043 | 20910048 | 5 | |
| Deletion | Chr7 | 24045888 | 24045891 | 3 | LOC_Os07g40050 |
| Deletion | Chr5 | 27393050 | 27393053 | 3 | |
| Deletion | Chr2 | 33055644 | 33055650 | 6 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 33502228 | 33502229 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 910487 | 1239386 | LOC_Os12g02660 |
| Inversion | Chr2 | 2139207 | 2151936 | LOC_Os02g04730 |
| Inversion | Chr2 | 2139244 | 2151959 | LOC_Os02g04730 |
| Inversion | Chr2 | 15635916 | 17026235 | 2 |
| Inversion | Chr4 | 19819990 | 19909091 | LOC_Os04g32820 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 751070 | Chr2 | 2691933 | |
| Translocation | Chr12 | 888354 | Chr5 | 15739828 | LOC_Os12g02570 |
| Translocation | Chr12 | 907459 | Chr2 | 15635956 | 2 |
| Translocation | Chr12 | 910477 | Chr2 | 17026235 | 2 |
| Translocation | Chr11 | 1003709 | Chr3 | 15239876 | LOC_Os03g26690 |
| Translocation | Chr11 | 1474966 | Chr9 | 2063963 | 2 |
| Translocation | Chr6 | 5893165 | Chr2 | 17306458 | LOC_Os06g11230 |
| Translocation | Chr10 | 14272468 | Chr1 | 9224470 | LOC_Os10g27080 |
| Translocation | Chr10 | 14272470 | Chr1 | 9223867 | LOC_Os10g27080 |
| Translocation | Chr10 | 18628327 | Chr5 | 9638215 | |
| Translocation | Chr4 | 34006449 | Chr3 | 18216866 | LOC_Os03g31839 |
