Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN904-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN904-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 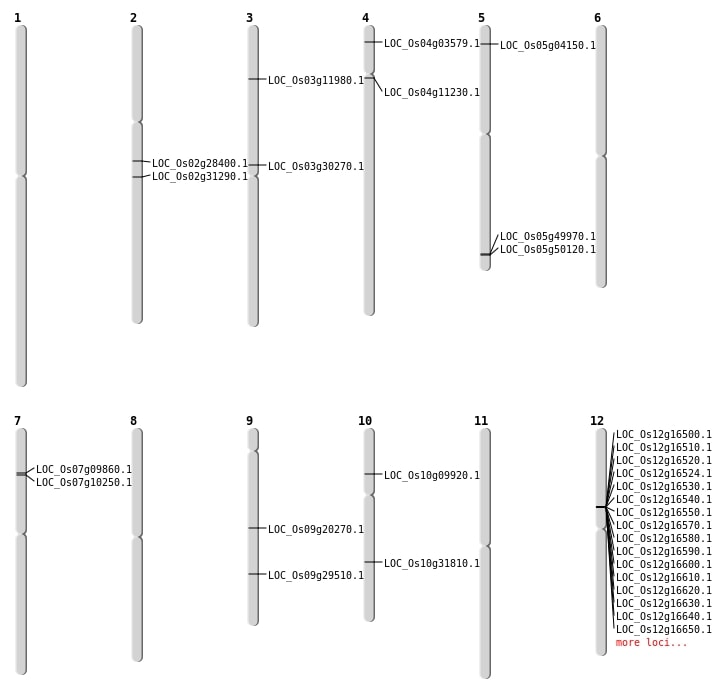
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15335726 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 16640000 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 40631854 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 40872185 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 15331023 | G-A | HOMO | INTRON | |
| SBS | Chr10 | 15331040 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 8661493 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 19226427 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 2045432 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 12277195 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 10524337 | C-A | HET | INTRON | |
| SBS | Chr2 | 6071945 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 12163501 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 12550076 | T-A | HET | INTRON | |
| SBS | Chr3 | 13116834 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 11676525 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 22379155 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 27649576 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 13875954 | A-T | HET | INTRON | |
| SBS | Chr5 | 16326650 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 25086131 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 28656904 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g49970 |
| SBS | Chr6 | 12115387 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 2541110 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 7043590 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 25927951 | T-G | HET | INTRON | |
| SBS | Chr8 | 11126563 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 402397 | A-G | HET | INTERGENIC |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1571843 | 1571850 | 7 | LOC_Os04g03579 |
| Deletion | Chr10 | 5369237 | 5369238 | 1 | LOC_Os10g09920 |
| Deletion | Chr3 | 6282479 | 6282485 | 6 | LOC_Os03g11980 |
| Deletion | Chr8 | 8050365 | 8050373 | 8 | |
| Deletion | Chr12 | 9448001 | 11030000 | 1582000 | 238 |
| Deletion | Chr12 | 10837491 | 10837498 | 7 | |
| Deletion | Chr1 | 13570362 | 13570364 | 2 | |
| Deletion | Chr12 | 14451689 | 14451690 | 1 | |
| Deletion | Chr5 | 15533964 | 15533972 | 8 | |
| Deletion | Chr10 | 15949190 | 15949195 | 5 | |
| Deletion | Chr10 | 16677842 | 16677843 | 1 | LOC_Os10g31810 |
| Deletion | Chr9 | 17951057 | 17951058 | 1 | LOC_Os09g29510 |
| Deletion | Chr2 | 18775767 | 18775768 | 1 | LOC_Os02g31290 |
| Deletion | Chr1 | 31435365 | 31435366 | 1 |
No Insertion
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 1862954 | 1921783 | LOC_Os05g04150 |
| Inversion | Chr5 | 1862980 | 1921793 | LOC_Os05g04150 |
| Inversion | Chr7 | 5245084 | 5505600 | 2 |
| Inversion | Chr11 | 9134393 | 27265402 | |
| Inversion | Chr4 | 29633855 | 29645943 |
Translocations: 14
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2994164 | Chr4 | 20494482 | |
| Translocation | Chr8 | 4115815 | Chr7 | 19337113 | |
| Translocation | Chr7 | 5505613 | Chr5 | 1862979 | 2 |
| Translocation | Chr7 | 5505844 | Chr5 | 1921792 | LOC_Os07g10250 |
| Translocation | Chr12 | 5600329 | Chr10 | 7819420 | |
| Translocation | Chr4 | 6139713 | Chr1 | 17190491 | LOC_Os04g11230 |
| Translocation | Chr9 | 6280172 | Chr8 | 18146151 | |
| Translocation | Chr6 | 8086714 | Chr5 | 5898797 | |
| Translocation | Chr4 | 8816658 | Chr3 | 21440252 | |
| Translocation | Chr9 | 12160255 | Chr7 | 5516692 | LOC_Os09g20270 |
| Translocation | Chr10 | 13618676 | Chr2 | 20514615 | |
| Translocation | Chr3 | 17277777 | Chr2 | 16784742 | 2 |
| Translocation | Chr12 | 23183547 | Chr6 | 23504958 | LOC_Os12g37760 |
| Translocation | Chr5 | 28724667 | Chr3 | 9391697 | LOC_Os05g50120 |
