Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN909-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN909-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 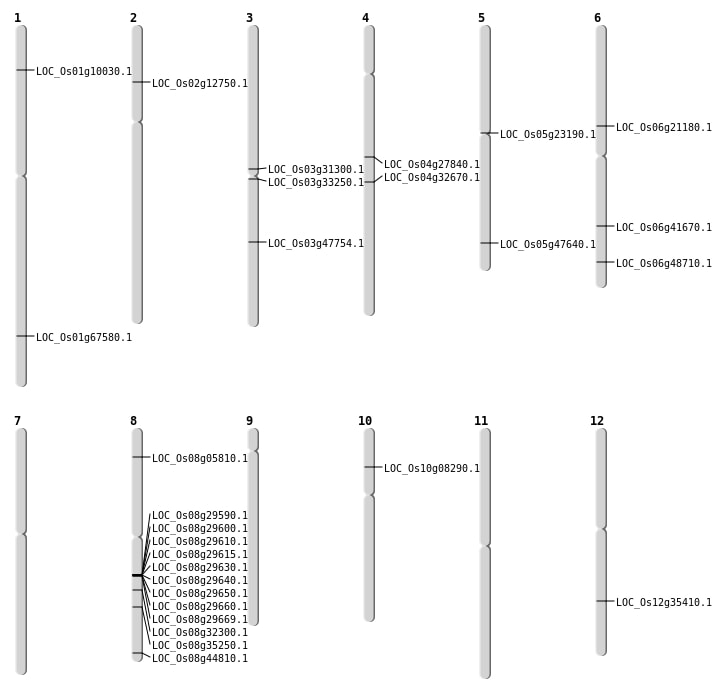
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16454432 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 17414983 | T-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 39278443 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g67580 |
| SBS | Chr1 | 40682834 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 6289971 | A-T | HET | INTRON | |
| SBS | Chr1 | 7547040 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr1 | 7834258 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 15664255 | T-C | HET | INTRON | |
| SBS | Chr10 | 5874190 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 6595022 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 8309036 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 17840071 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 17840072 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 6187970 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 34410970 | C-G | HOMO | INTRON | |
| SBS | Chr2 | 5981645 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 6677207 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g12750 |
| SBS | Chr3 | 15746664 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 26938162 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 27096791 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g47754 |
| SBS | Chr3 | 27939509 | C-T | HET | INTRON | |
| SBS | Chr3 | 7977708 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 21879189 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 13225335 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g23190 |
| SBS | Chr6 | 12242615 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g21180 |
| SBS | Chr6 | 29468396 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g48710 |
| SBS | Chr6 | 7064608 | C-T | HET | INTRON | |
| SBS | Chr7 | 17605640 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 15473392 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 15937045 | G-A | HET | INTRON | |
| SBS | Chr8 | 18997007 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 22242183 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g35250 |
| SBS | Chr8 | 28141586 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g44810 |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 3116651 | 3116655 | 4 | LOC_Os08g05810 |
| Deletion | Chr12 | 4718910 | 4718911 | 1 | |
| Deletion | Chr3 | 5187222 | 5187233 | 11 | |
| Deletion | Chr6 | 5910639 | 5910640 | 1 | |
| Deletion | Chr3 | 7308624 | 7308625 | 1 | |
| Deletion | Chr3 | 7479713 | 7479714 | 1 | |
| Deletion | Chr3 | 8115060 | 8115062 | 2 | |
| Deletion | Chr2 | 8126695 | 8126703 | 8 | |
| Deletion | Chr5 | 8933043 | 8933067 | 24 | |
| Deletion | Chr1 | 11395795 | 11395804 | 9 | |
| Deletion | Chr10 | 14131850 | 14131852 | 2 | |
| Deletion | Chr10 | 15464990 | 15464992 | 2 | |
| Deletion | Chr2 | 15887100 | 15887102 | 2 | |
| Deletion | Chr4 | 15962746 | 15962755 | 9 | |
| Deletion | Chr4 | 16447514 | 16448154 | 640 | LOC_Os04g27840 |
| Deletion | Chr2 | 17182648 | 17182654 | 6 | |
| Deletion | Chr1 | 17802644 | 17802656 | 12 | |
| Deletion | Chr8 | 18148001 | 18223000 | 75000 | 9 |
| Deletion | Chr3 | 19022773 | 19022775 | 2 | LOC_Os03g33250 |
| Deletion | Chr9 | 20029017 | 20029021 | 4 | |
| Deletion | Chr3 | 20165359 | 20165378 | 19 | |
| Deletion | Chr3 | 21814091 | 21814095 | 4 | |
| Deletion | Chr12 | 24236399 | 24236415 | 16 | |
| Deletion | Chr6 | 24955313 | 24955316 | 3 | LOC_Os06g41670 |
| Deletion | Chr3 | 26960470 | 26960493 | 23 | |
| Deletion | Chr12 | 27277306 | 27277308 | 2 | |
| Deletion | Chr5 | 27292465 | 27292475 | 10 | LOC_Os05g47640 |
| Deletion | Chr1 | 27414361 | 27414362 | 1 | |
| Deletion | Chr1 | 38241934 | 38241940 | 6 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 8211430 | 8211430 | 1 | |
| Insertion | Chr12 | 1185381 | 1185381 | 1 | |
| Insertion | Chr3 | 17832414 | 17832420 | 7 | LOC_Os03g31300 |
| Insertion | Chr4 | 19690674 | 19690675 | 2 | LOC_Os04g32670 |
| Insertion | Chr4 | 21130579 | 21130580 | 2 | |
| Insertion | Chr5 | 11628116 | 11628116 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 18144177 | 20009579 | LOC_Os08g32300 |
| Inversion | Chr8 | 18223067 | 20009595 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 5678755 | Chr1 | 22198164 | |
| Translocation | Chr10 | 6595051 | Chr4 | 7489899 | |
| Translocation | Chr12 | 12883304 | Chr10 | 4480981 | LOC_Os10g08290 |
| Translocation | Chr11 | 14627735 | Chr1 | 5228946 | LOC_Os01g10030 |
| Translocation | Chr12 | 21538364 | Chr8 | 18146558 | LOC_Os12g35410 |
| Translocation | Chr3 | 22942505 | Chr1 | 39679150 | |
| Translocation | Chr11 | 27253702 | Chr7 | 22207938 |
