Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN910-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN910-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 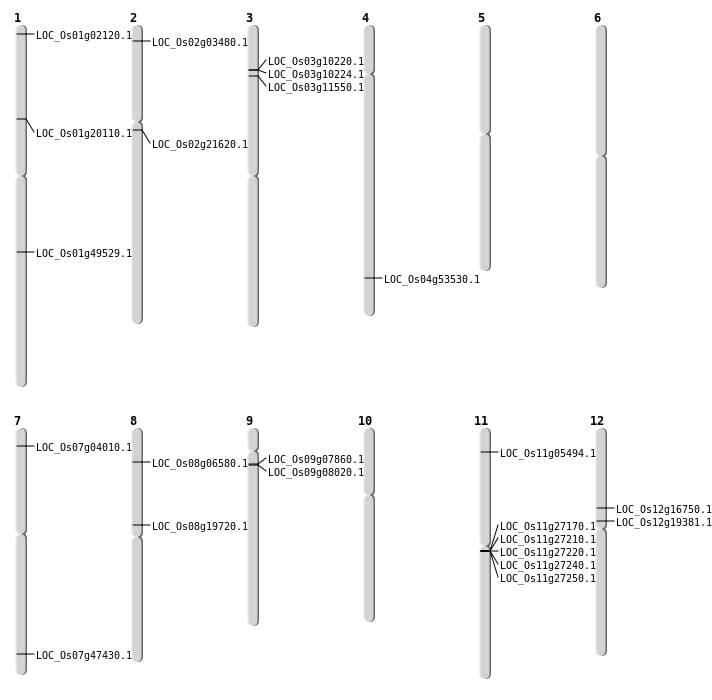
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24292437 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 24838306 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 27168784 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 27206905 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 3624519 | T-G | HET | INTRON | |
| SBS | Chr1 | 36846060 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 51007 | G-A | HET | INTRON | |
| SBS | Chr10 | 10981069 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1240199 | T-A | HOMO | INTRON | |
| SBS | Chr11 | 15681189 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 24294838 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 19898385 | A-C | HET | INTRON | |
| SBS | Chr12 | 22831884 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 12840828 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g21620 |
| SBS | Chr2 | 3760275 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 15761086 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 30157618 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 3039189 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 5683474 | A-T | HET | INTRON | |
| SBS | Chr3 | 9791623 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 22841857 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 23205912 | A-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 27415407 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 12804950 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 13380304 | A-T | HOMO | INTRON | |
| SBS | Chr5 | 18643766 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 19933504 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 24297705 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 25325367 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 5069510 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 1694018 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g04010 |
| SBS | Chr7 | 26844200 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 8563563 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 8914091 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 24826593 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 3717483 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g06580 |
| SBS | Chr9 | 10383447 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 3997712 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g07860 |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 614939 | 615053 | 114 | LOC_Os01g02120 |
| Deletion | Chr6 | 2391194 | 2391196 | 2 | |
| Deletion | Chr9 | 4130896 | 4130897 | 1 | LOC_Os09g08020 |
| Deletion | Chr12 | 4785173 | 4785179 | 6 | |
| Deletion | Chr3 | 5204260 | 5204269 | 9 | 2 |
| Deletion | Chr3 | 5983862 | 5983880 | 18 | LOC_Os03g11550 |
| Deletion | Chr5 | 6072765 | 6072766 | 1 | |
| Deletion | Chr6 | 6665371 | 6665373 | 2 | |
| Deletion | Chr7 | 6816026 | 6816035 | 9 | |
| Deletion | Chr7 | 6911659 | 6911665 | 6 | |
| Deletion | Chr10 | 7680564 | 7680566 | 2 | |
| Deletion | Chr4 | 8116440 | 8116441 | 1 | |
| Deletion | Chr1 | 8998673 | 8998691 | 18 | |
| Deletion | Chr12 | 9603238 | 9603248 | 10 | LOC_Os12g16750 |
| Deletion | Chr3 | 9743265 | 9743269 | 4 | |
| Deletion | Chr9 | 10383179 | 10383238 | 59 | |
| Deletion | Chr1 | 11412145 | 11412162 | 17 | LOC_Os01g20110 |
| Deletion | Chr8 | 11808772 | 11808775 | 3 | LOC_Os08g19720 |
| Deletion | Chr7 | 11862133 | 11862165 | 32 | |
| Deletion | Chr6 | 14029046 | 14029073 | 27 | |
| Deletion | Chr8 | 15234644 | 15234655 | 11 | |
| Deletion | Chr11 | 15646001 | 15679000 | 33000 | 5 |
| Deletion | Chr3 | 17922323 | 17922330 | 7 | |
| Deletion | Chr9 | 18966903 | 18966916 | 13 | |
| Deletion | Chr4 | 19425335 | 19425339 | 4 | |
| Deletion | Chr5 | 20636861 | 20636874 | 13 | |
| Deletion | Chr4 | 21888030 | 21888040 | 10 | |
| Deletion | Chr5 | 21944336 | 21944352 | 16 | |
| Deletion | Chr7 | 23943580 | 23943592 | 12 | |
| Deletion | Chr4 | 28336442 | 28336447 | 5 | |
| Deletion | Chr7 | 28359990 | 28359998 | 8 | LOC_Os07g47430 |
| Deletion | Chr1 | 28486676 | 28486683 | 7 | LOC_Os01g49529 |
| Deletion | Chr4 | 31895323 | 31895324 | 1 | LOC_Os04g53530 |
| Deletion | Chr1 | 33024943 | 33024980 | 37 | |
| Deletion | Chr1 | 36080155 | 36080183 | 28 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 17838567 | 17838570 | 4 | |
| Insertion | Chr2 | 12353499 | 12353502 | 4 | |
| Insertion | Chr2 | 1417698 | 1417699 | 2 | LOC_Os02g03480 |
| Insertion | Chr3 | 381152 | 381157 | 6 | |
| Insertion | Chr4 | 16564311 | 16564332 | 22 | |
| Insertion | Chr4 | 32487617 | 32487618 | 2 | |
| Insertion | Chr4 | 7471318 | 7471318 | 1 | |
| Insertion | Chr7 | 12851170 | 12851171 | 2 | |
| Insertion | Chr7 | 3165265 | 3165266 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 15645671 | 15900018 | LOC_Os11g27170 |
| Inversion | Chr11 | 15679006 | 15900034 | LOC_Os11g27250 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 2482047 | Chr1 | 614953 | 2 |
| Translocation | Chr12 | 11272894 | Chr7 | 2280249 | LOC_Os12g19381 |
| Translocation | Chr12 | 18596792 | Chr5 | 2015452 | |
| Translocation | Chr11 | 21145101 | Chr8 | 26805614 | |
| Translocation | Chr4 | 27473358 | Chr3 | 13639296 |
