Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN915-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN915-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 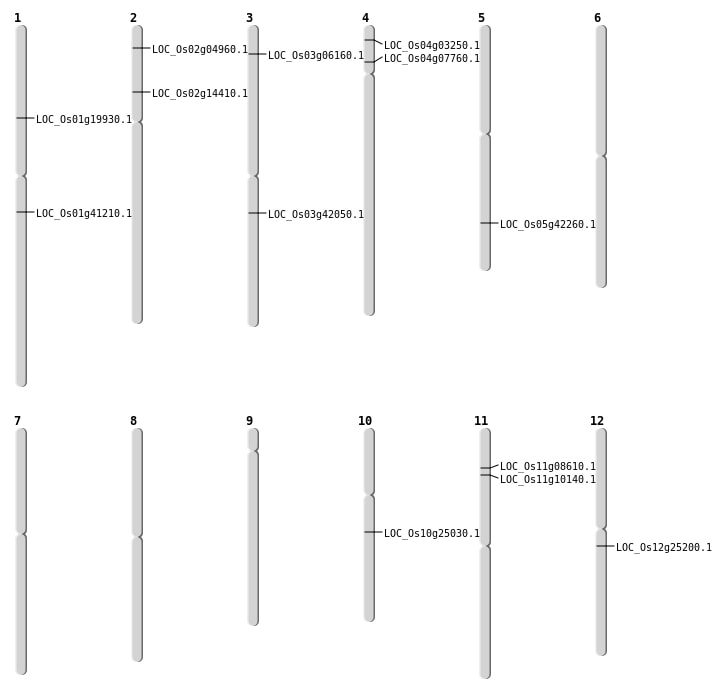
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10524050 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 11313473 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g19930 |
| SBS | Chr1 | 20738081 | C-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 29348305 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 3546018 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 42812911 | G-A | HET | INTRON | |
| SBS | Chr10 | 16858722 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 5020567 | C-T | HET | INTRON | |
| SBS | Chr10 | 5095032 | A-G | HET | INTRON | |
| SBS | Chr11 | 19906420 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 4579083 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g08610 |
| SBS | Chr11 | 5492881 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g10140 |
| SBS | Chr12 | 11040530 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 12105767 | T-A | HET | INTRON | |
| SBS | Chr12 | 14489385 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g25200 |
| SBS | Chr2 | 10155341 | T-C | HOMO | UTR_5_PRIME | |
| SBS | Chr2 | 29333375 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 23374544 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g42050 |
| SBS | Chr3 | 25002069 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 3092144 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g06160 |
| SBS | Chr3 | 33330370 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 4467224 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 33511067 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 33847085 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 9068292 | C-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 11218508 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 15374966 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 24721102 | C-A | HET | INTRON | |
| SBS | Chr5 | 24721105 | A-C | HET | INTRON | |
| SBS | Chr6 | 13004498 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 15322413 | C-T | HET | INTRON | |
| SBS | Chr7 | 16536956 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 22328991 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 21307670 | G-T | HET | INTRON | |
| SBS | Chr8 | 4881368 | G-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 6426513 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 6443438 | G-T | HET | INTRON | |
| SBS | Chr8 | 7565437 | T-A | HOMO | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 2327247 | 2327258 | 11 | LOC_Os02g04960 |
| Deletion | Chr8 | 4195367 | 4195369 | 2 | |
| Deletion | Chr6 | 6212623 | 6212634 | 11 | |
| Deletion | Chr8 | 7349850 | 7349852 | 2 | |
| Deletion | Chr1 | 7906001 | 7916000 | 10000 | |
| Deletion | Chr5 | 7922403 | 7922405 | 2 | |
| Deletion | Chr2 | 10162399 | 10162403 | 4 | |
| Deletion | Chr7 | 10571429 | 10571434 | 5 | |
| Deletion | Chr5 | 13243713 | 13243726 | 13 | |
| Deletion | Chr7 | 15506662 | 15506677 | 15 | |
| Deletion | Chr10 | 15608523 | 15608524 | 1 | |
| Deletion | Chr5 | 15766429 | 15766463 | 34 | |
| Deletion | Chr10 | 20179281 | 20179292 | 11 | |
| Deletion | Chr8 | 21696697 | 21696714 | 17 | |
| Deletion | Chr2 | 22914338 | 22914442 | 104 | |
| Deletion | Chr5 | 24720888 | 24720889 | 1 | LOC_Os05g42260 |
| Deletion | Chr3 | 28103223 | 28103237 | 14 | |
| Deletion | Chr7 | 28169870 | 28169871 | 1 | |
| Deletion | Chr7 | 28606179 | 28606180 | 1 | |
| Deletion | Chr3 | 30852330 | 30852332 | 2 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23331772 | 23331817 | 46 | LOC_Os01g41210 |
| Insertion | Chr1 | 4807434 | 4807435 | 2 | |
| Insertion | Chr12 | 11007901 | 11007902 | 2 | |
| Insertion | Chr12 | 3603698 | 3603702 | 5 | |
| Insertion | Chr4 | 4124944 | 4125002 | 59 | LOC_Os04g07760 |
| Insertion | Chr5 | 10133374 | 10133375 | 2 | |
| Insertion | Chr5 | 7365322 | 7365323 | 2 | |
| Insertion | Chr6 | 24037839 | 24037842 | 4 | |
| Insertion | Chr8 | 11243534 | 11243535 | 2 | |
| Insertion | Chr8 | 26815767 | 26815768 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 5785984 | 7318438 | |
| Inversion | Chr10 | 10438107 | 10543747 | |
| Inversion | Chr10 | 12886954 | 14239730 | LOC_Os10g25030 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 1384769 | Chr2 | 7913909 | 2 |
| Translocation | Chr12 | 4980276 | Chr3 | 15238097 | |
| Translocation | Chr8 | 7485347 | Chr7 | 12529689 | |
| Translocation | Chr11 | 12004224 | Chr10 | 2305634 | |
| Translocation | Chr11 | 15905659 | Chr4 | 22046396 | |
| Translocation | Chr11 | 27773718 | Chr5 | 2205885 |
