Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN924-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN924-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 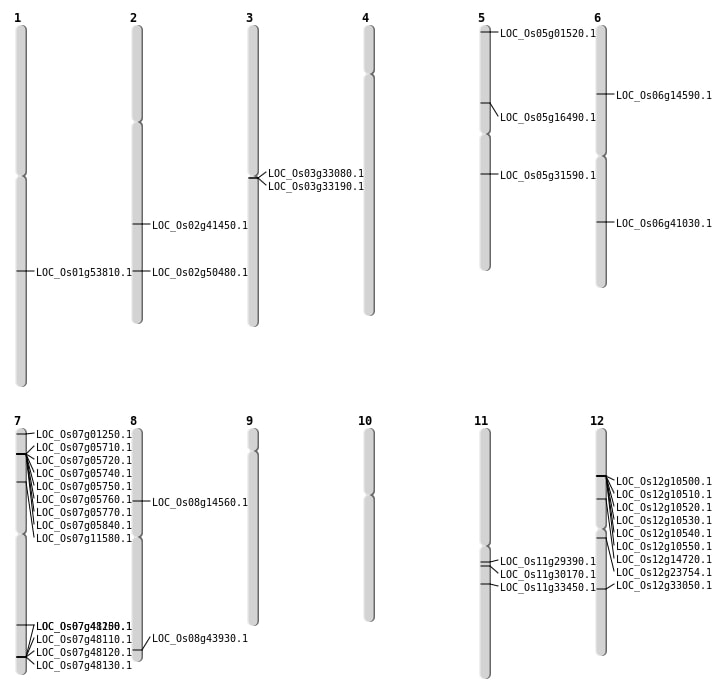
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 241341 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 29344350 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 30934670 | G-T | HET | STOP_GAINED | LOC_Os01g53810 |
| SBS | Chr1 | 30934671 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g53810 |
| SBS | Chr10 | 5783411 | T-A | HET | INTRON | |
| SBS | Chr11 | 17538111 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g30170 |
| SBS | Chr11 | 23287750 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 24247251 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 577918 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 16315406 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 11077561 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 16000187 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 31310634 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 487194 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 6763695 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 9996929 | G-T | HOMO | INTRON | |
| SBS | Chr3 | 18817260 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 20549523 | A-T | HET | INTRON | |
| SBS | Chr4 | 15514881 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21215831 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr5 | 21299737 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 21784523 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 24057352 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 4938222 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 9358372 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g16490 |
| SBS | Chr6 | 11507766 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 22169286 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 8212931 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g14590 |
| SBS | Chr8 | 5194750 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 8752881 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g14560 |
| SBS | Chr8 | 9753069 | G-A | HET | INTRON | |
| SBS | Chr9 | 5235240 | C-A | HET | INTRON |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 2724001 | 2781000 | 57000 | 6 |
| Deletion | Chr7 | 2807218 | 2807227 | 9 | LOC_Os07g05840 |
| Deletion | Chr12 | 5561001 | 5598000 | 37000 | 6 |
| Deletion | Chr7 | 6424449 | 6424456 | 7 | |
| Deletion | Chr10 | 8954130 | 8954141 | 11 | |
| Deletion | Chr2 | 11554902 | 11554903 | 1 | |
| Deletion | Chr10 | 11939146 | 11939160 | 14 | |
| Deletion | Chr12 | 13487392 | 13487394 | 2 | LOC_Os12g23754 |
| Deletion | Chr9 | 14958223 | 14958235 | 12 | |
| Deletion | Chr9 | 18330635 | 18330653 | 18 | |
| Deletion | Chr9 | 19790871 | 19790877 | 6 | |
| Deletion | Chr8 | 20632066 | 20632072 | 6 | |
| Deletion | Chr6 | 21089331 | 21089336 | 5 | |
| Deletion | Chr6 | 21239398 | 21239407 | 9 | |
| Deletion | Chr6 | 24507328 | 24507329 | 1 | LOC_Os06g41030 |
| Deletion | Chr7 | 24721925 | 24721928 | 3 | LOC_Os07g41250 |
| Deletion | Chr8 | 27683322 | 27683328 | 6 | LOC_Os08g43930 |
| Deletion | Chr6 | 28454187 | 28454190 | 3 | |
| Deletion | Chr7 | 28727013 | 28747879 | 20866 | 4 |
| Deletion | Chr4 | 28997824 | 28997828 | 4 | |
| Deletion | Chr3 | 29635276 | 29635288 | 12 | |
| Deletion | Chr2 | 30825789 | 30825790 | 1 | LOC_Os02g50480 |
| Deletion | Chr1 | 39535984 | 39535985 | 1 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 42459782 | 42459783 | 2 | |
| Insertion | Chr2 | 12864063 | 12864066 | 4 | |
| Insertion | Chr2 | 20545991 | 20545992 | 2 | |
| Insertion | Chr4 | 4266037 | 4266038 | 2 | |
| Insertion | Chr4 | 4664657 | 4664657 | 1 | |
| Insertion | Chr5 | 311092 | 311093 | 2 | LOC_Os05g01520 |
| Insertion | Chr8 | 9184146 | 9184148 | 3 | |
| Insertion | Chr9 | 13194256 | 13194256 | 1 | |
| Insertion | Chr9 | 7196896 | 7196897 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 149645 | 2494166 | LOC_Os07g01250 |
| Inversion | Chr12 | 8246555 | 8782149 | |
| Inversion | Chr12 | 8425013 | 8782144 | LOC_Os12g14720 |
| Inversion | Chr3 | 18927701 | 18987801 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 689428 | Chr2 | 24817280 | LOC_Os02g41450 |
| Translocation | Chr9 | 1518271 | Chr5 | 18390626 | LOC_Os05g31590 |
| Translocation | Chr9 | 6318970 | Chr1 | 1281619 | |
| Translocation | Chr11 | 7190216 | Chr1 | 14546653 | |
| Translocation | Chr8 | 8400857 | Chr7 | 6388311 | LOC_Os07g11580 |
| Translocation | Chr8 | 12181098 | Chr6 | 14299325 | |
| Translocation | Chr11 | 19804903 | Chr3 | 23443050 | LOC_Os11g33450 |
| Translocation | Chr12 | 19969409 | Chr11 | 17059952 | 2 |
