Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN927-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN927-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 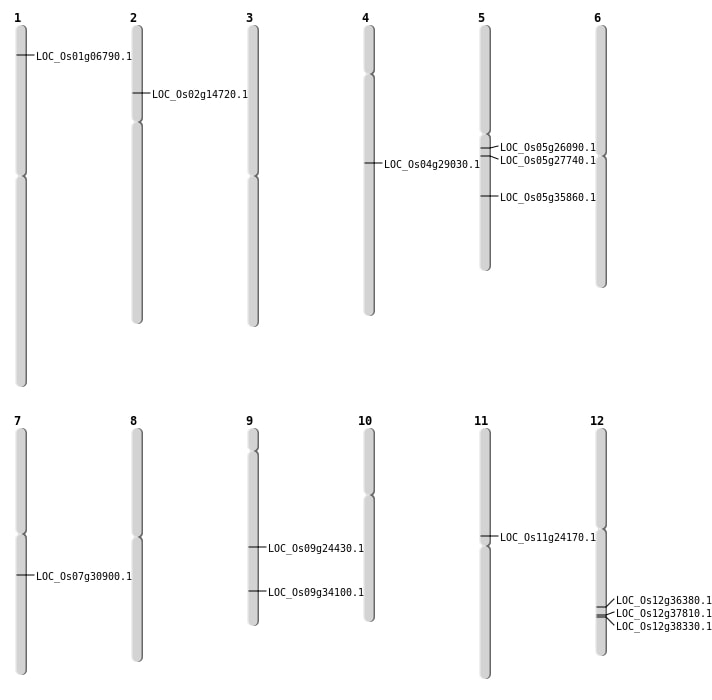
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17170657 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 27981589 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 35537305 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 6577898 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 22721786 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 3372207 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 9287344 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 11786701 | A-C | HET | INTRON | |
| SBS | Chr11 | 13752278 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 20620082 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 23995148 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 15788060 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 21426081 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 22272343 | T-G | HOMO | STOP_GAINED | LOC_Os12g36380 |
| SBS | Chr2 | 24660347 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 30370446 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 9256331 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 9256332 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 15463908 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 23457074 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 30705752 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 15526245 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 9584821 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 11925810 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 15172617 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g26090 |
| SBS | Chr5 | 18572048 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 18572053 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 21360760 | T-A | HET | INTRON | |
| SBS | Chr5 | 3400591 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 27655259 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 14025333 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 18290247 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30900 |
| SBS | Chr7 | 19055779 | A-C | HET | INTRON | |
| SBS | Chr8 | 15996787 | C-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 19992492 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 25677846 | C-T | HET | INTRON | |
| SBS | Chr8 | 4216060 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 13147300 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 14528624 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g24430 |
| SBS | Chr9 | 14528626 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g24430 |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 716007 | 716021 | 14 | |
| Deletion | Chr9 | 1744322 | 1744323 | 1 | |
| Deletion | Chr2 | 2059555 | 2059557 | 2 | |
| Deletion | Chr2 | 2180194 | 2180216 | 22 | |
| Deletion | Chr2 | 3023551 | 3023552 | 1 | |
| Deletion | Chr2 | 3748290 | 3748291 | 1 | |
| Deletion | Chr2 | 8163789 | 8163791 | 2 | |
| Deletion | Chr2 | 9247015 | 9247030 | 15 | |
| Deletion | Chr11 | 10278640 | 10278645 | 5 | |
| Deletion | Chr1 | 10389108 | 10389113 | 5 | |
| Deletion | Chr1 | 10706553 | 10706571 | 18 | |
| Deletion | Chr11 | 13747133 | 13747142 | 9 | LOC_Os11g24170 |
| Deletion | Chr4 | 17203345 | 17203352 | 7 | LOC_Os04g29030 |
| Deletion | Chr12 | 17575954 | 17575955 | 1 | |
| Deletion | Chr2 | 18875418 | 18875433 | 15 | |
| Deletion | Chr5 | 19063329 | 19063335 | 6 | |
| Deletion | Chr5 | 21277670 | 21277672 | 2 | LOC_Os05g35860 |
| Deletion | Chr11 | 21795264 | 21795265 | 1 | |
| Deletion | Chr5 | 24373217 | 24373239 | 22 | |
| Deletion | Chr5 | 28077027 | 28077038 | 11 | |
| Deletion | Chr1 | 31855071 | 31855078 | 7 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 34064088 | 34064089 | 2 | |
| Insertion | Chr2 | 17866036 | 17866037 | 2 | |
| Insertion | Chr3 | 8934064 | 8934064 | 1 | |
| Insertion | Chr9 | 10327470 | 10327471 | 2 |
Inversions: 5
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 60399 | Chr2 | 8120310 | LOC_Os02g14720 |
| Translocation | Chr6 | 8650813 | Chr4 | 8676809 | |
| Translocation | Chr2 | 10396555 | Chr1 | 15263540 | |
| Translocation | Chr9 | 12257796 | Chr1 | 3222415 | LOC_Os01g06790 |
| Translocation | Chr6 | 14457663 | Chr4 | 8094911 | |
| Translocation | Chr9 | 18223699 | Chr5 | 16159008 | LOC_Os05g27740 |
| Translocation | Chr9 | 20127285 | Chr4 | 5191228 | LOC_Os09g34100 |
| Translocation | Chr9 | 20793648 | Chr4 | 5192075 | |
| Translocation | Chr12 | 23524435 | Chr9 | 20793659 | LOC_Os12g38330 |
