Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN928-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN928-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 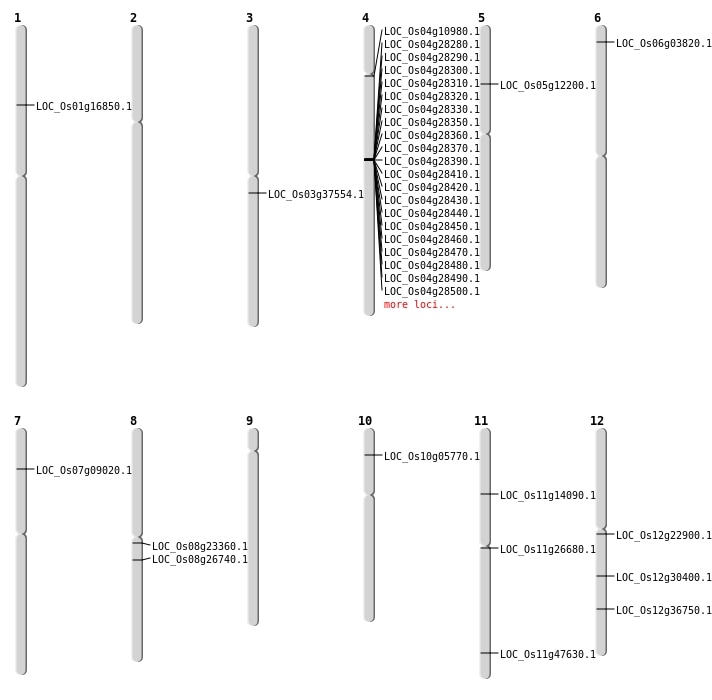
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11331011 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 22845381 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 23057070 | G-T | HET | INTRON | |
| SBS | Chr1 | 23263183 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 26664747 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 4293748 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 21400801 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 28766332 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g47630 |
| SBS | Chr11 | 9046155 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 12932499 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g22900 |
| SBS | Chr12 | 22365167 | G-C | HOMO | INTRON | |
| SBS | Chr12 | 22788234 | C-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 2777324 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 6266318 | T-A | HET | INTRON | |
| SBS | Chr2 | 9965888 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 31941773 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 10910929 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 15542454 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 17334314 | G-A | HET | INTRON | |
| SBS | Chr4 | 23381239 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 28967421 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 4875277 | A-C | HET | UTR_5_PRIME | |
| SBS | Chr4 | 5580144 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 20835774 | G-A | HET | INTRON | |
| SBS | Chr5 | 23133274 | G-A | HET | INTRON | |
| SBS | Chr5 | 6987616 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g12200 |
| SBS | Chr5 | 8976316 | A-C | HET | INTRON | |
| SBS | Chr6 | 1228421 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 1531158 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g03820 |
| SBS | Chr6 | 1531159 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g03820 |
| SBS | Chr6 | 29049402 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 1558613 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 21784728 | T-A | HET | INTRON | |
| SBS | Chr7 | 21784729 | G-A | HET | INTRON | |
| SBS | Chr7 | 21784730 | T-A | HET | INTRON | |
| SBS | Chr7 | 27081901 | A-C | HET | INTRON | |
| SBS | Chr7 | 28318798 | G-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 11002016 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 17598255 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr8 | 9601011 | G-A | HOMO | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2263966 | 2263968 | 2 | |
| Deletion | Chr8 | 2818405 | 2818411 | 6 | |
| Deletion | Chr10 | 2851482 | 2851483 | 1 | |
| Deletion | Chr7 | 4708855 | 4708869 | 14 | LOC_Os07g09020 |
| Deletion | Chr1 | 5589558 | 5589560 | 2 | |
| Deletion | Chr4 | 5960078 | 5960081 | 3 | LOC_Os04g10980 |
| Deletion | Chr11 | 7845315 | 7845319 | 4 | LOC_Os11g14090 |
| Deletion | Chr11 | 7891333 | 7891335 | 2 | |
| Deletion | Chr7 | 8887283 | 8887287 | 4 | |
| Deletion | Chr5 | 11006717 | 11006722 | 5 | |
| Deletion | Chr10 | 11508156 | 11508487 | 331 | |
| Deletion | Chr11 | 15283402 | 15283414 | 12 | LOC_Os11g26680 |
| Deletion | Chr12 | 16069616 | 16069617 | 1 | |
| Deletion | Chr4 | 16717001 | 16776000 | 59000 | 7 |
| Deletion | Chr4 | 16778001 | 17140000 | 362000 | 54 |
| Deletion | Chr4 | 17151001 | 17158000 | 7000 | 2 |
| Deletion | Chr12 | 18245132 | 18245135 | 3 | LOC_Os12g30400 |
| Deletion | Chr10 | 18411080 | 18411081 | 1 | |
| Deletion | Chr5 | 20054388 | 20054397 | 9 | |
| Deletion | Chr7 | 21809926 | 21809931 | 5 | |
| Deletion | Chr12 | 22385147 | 22385157 | 10 | |
| Deletion | Chr12 | 22510959 | 22512225 | 1266 | LOC_Os12g36750 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 27699077 | 27699078 | 2 | |
| Insertion | Chr11 | 9940605 | 9940608 | 4 | |
| Insertion | Chr9 | 6713580 | 6713581 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 9616400 | 14360029 | LOC_Os01g16850 |
| Inversion | Chr8 | 16161801 | 16266407 | LOC_Os08g26740 |
| Inversion | Chr1 | 16912125 | 16945212 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2906555 | Chr5 | 26269524 | LOC_Os10g05770 |
| Translocation | Chr11 | 8558337 | Chr4 | 16768817 | |
| Translocation | Chr8 | 14115861 | Chr3 | 20826959 | 2 |
| Translocation | Chr8 | 14117320 | Chr3 | 20826893 | 2 |
| Translocation | Chr11 | 25275701 | Chr2 | 9161375 |
