Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN934-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN934-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 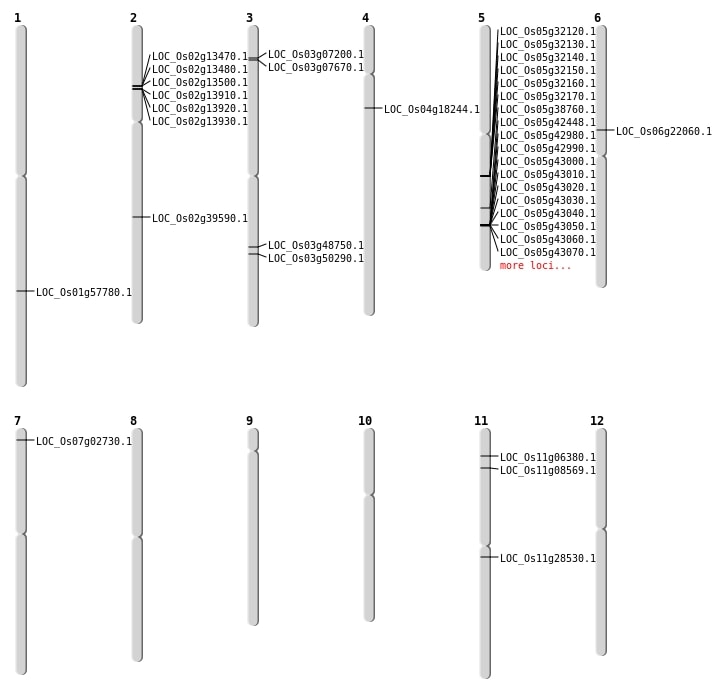
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 28463737 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 42773147 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 12080360 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 9536299 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 23893405 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 23893406 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g39590 |
| SBS | Chr2 | 6932660 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 7669450 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 180865 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 23173314 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 27779917 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g48750 |
| SBS | Chr3 | 5024961 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10086870 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18244 |
| SBS | Chr4 | 149923 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 16160763 | A-G | HET | INTRON | |
| SBS | Chr5 | 10229119 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 15960416 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 18724150 | C-G | HET | INTRON | |
| SBS | Chr5 | 22744155 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g38760 |
| SBS | Chr5 | 23206183 | T-C | HET | INTRON | |
| SBS | Chr5 | 26343482 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 29093298 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 12797715 | C-A | HOMO | STOP_GAINED | LOC_Os06g22060 |
| SBS | Chr6 | 14273911 | G-C | HOMO | INTRON | |
| SBS | Chr6 | 17899238 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 22087969 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 3717452 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 17248309 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 20190143 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 17113442 | A-C | HOMO | INTRON | |
| SBS | Chr8 | 25820486 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 5820529 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 6958791 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 15572200 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 2629915 | C-T | HOMO | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 659540 | 659542 | 2 | |
| Deletion | Chr7 | 993576 | 994368 | 792 | LOC_Os07g02730 |
| Deletion | Chr8 | 2607364 | 2607365 | 1 | |
| Deletion | Chr8 | 3302936 | 3302946 | 10 | |
| Deletion | Chr2 | 7188001 | 7204000 | 16000 | 2 |
| Deletion | Chr2 | 7568264 | 7584839 | 16575 | 3 |
| Deletion | Chr5 | 8292464 | 8292466 | 2 | |
| Deletion | Chr1 | 10016673 | 10016674 | 1 | |
| Deletion | Chr1 | 10369573 | 10369631 | 58 | |
| Deletion | Chr4 | 12404917 | 12404931 | 14 | |
| Deletion | Chr2 | 12695257 | 12697974 | 2717 | |
| Deletion | Chr1 | 16069119 | 16069123 | 4 | |
| Deletion | Chr4 | 17717308 | 17717311 | 3 | |
| Deletion | Chr5 | 18728419 | 18761578 | 33159 | 6 |
| Deletion | Chr11 | 21064606 | 21064623 | 17 | |
| Deletion | Chr11 | 21518397 | 21518398 | 1 | |
| Deletion | Chr3 | 21706066 | 21706090 | 24 | |
| Deletion | Chr3 | 22246659 | 22246660 | 1 | |
| Deletion | Chr5 | 22834668 | 22834721 | 53 | |
| Deletion | Chr5 | 24922087 | 24922088 | 1 | LOC_Os05g42448 |
| Deletion | Chr5 | 24959001 | 25055000 | 96000 | 18 |
| Deletion | Chr3 | 28183930 | 28183935 | 5 | |
| Deletion | Chr3 | 28663891 | 28663892 | 1 | LOC_Os03g50290 |
| Deletion | Chr1 | 33419785 | 33419786 | 1 | LOC_Os01g57780 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 3253467 | 3253467 | 1 | |
| Insertion | Chr10 | 2306962 | 2306973 | 12 | |
| Insertion | Chr12 | 24700868 | 24700868 | 1 | |
| Insertion | Chr6 | 23644970 | 23644990 | 21 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 3071508 | 4545754 | 2 |
| Inversion | Chr3 | 3684353 | 3895402 | 2 |
| Inversion | Chr2 | 6213777 | 7702982 | |
| Inversion | Chr2 | 7204316 | 7905462 | LOC_Os02g13480 |
| Inversion | Chr2 | 7215506 | 12697974 | LOC_Os02g13500 |
| Inversion | Chr2 | 7215520 | 12695261 | LOC_Os02g13500 |
| Inversion | Chr5 | 18728420 | 18877658 | |
| Inversion | Chr4 | 20047186 | 20060232 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 473794 | Chr1 | 24174134 | |
| Translocation | Chr10 | 1823325 | Chr1 | 34016451 | |
| Translocation | Chr11 | 16435523 | Chr5 | 18740007 | 2 |
