Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN941-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN941-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 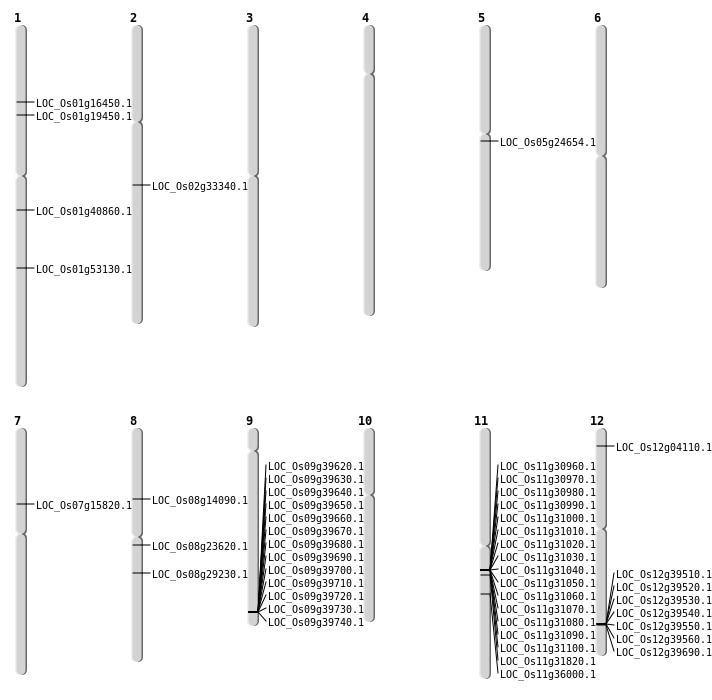
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 28959188 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 29474288 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 13241432 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 16024173 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 18124939 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 18693052 | T-A | HET | INTRON | |
| SBS | Chr11 | 23757903 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 24790388 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 10114273 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 10547529 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 1720970 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g04110 |
| SBS | Chr12 | 22637637 | C-T | HET | INTRON | |
| SBS | Chr12 | 23512677 | C-T | HET | INTRON | |
| SBS | Chr2 | 27283146 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 5110046 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 34105201 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 31180642 | T-A | HET | INTRON | |
| SBS | Chr5 | 26904105 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 27200736 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 29511934 | T-A | HOMO | UTR_5_PRIME | |
| SBS | Chr5 | 29511936 | G-A | HOMO | UTR_5_PRIME | |
| SBS | Chr5 | 5747247 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 8241039 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 30497465 | T-A | HET | INTRON | |
| SBS | Chr6 | 6635393 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19377178 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 2022858 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 9189255 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 9189257 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g15820 |
| SBS | Chr7 | 9189259 | C-A | HOMO | START_LOST | LOC_Os07g15820 |
| SBS | Chr8 | 18323858 | A-C | HOMO | INTRON | |
| SBS | Chr8 | 23097504 | C-T | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1873551 | 1873555 | 4 | |
| Deletion | Chr12 | 5334289 | 5334296 | 7 | |
| Deletion | Chr1 | 11005933 | 11005938 | 5 | LOC_Os01g19450 |
| Deletion | Chr1 | 12264720 | 12264721 | 1 | |
| Deletion | Chr5 | 14255029 | 14255036 | 7 | LOC_Os05g24654 |
| Deletion | Chr10 | 14664502 | 14664504 | 2 | |
| Deletion | Chr5 | 15822849 | 15822850 | 1 | |
| Deletion | Chr12 | 16728177 | 16728178 | 1 | |
| Deletion | Chr8 | 17924301 | 17924323 | 22 | LOC_Os08g29230 |
| Deletion | Chr11 | 18015001 | 18097000 | 82000 | 15 |
| Deletion | Chr11 | 18692087 | 18692959 | 872 | LOC_Os11g31820 |
| Deletion | Chr2 | 19810522 | 19810525 | 3 | LOC_Os02g33340 |
| Deletion | Chr8 | 20234879 | 20234920 | 41 | |
| Deletion | Chr11 | 21156959 | 21156960 | 1 | LOC_Os11g36000 |
| Deletion | Chr12 | 21298864 | 21298879 | 15 | |
| Deletion | Chr10 | 21949048 | 21949077 | 29 | |
| Deletion | Chr5 | 22324440 | 22324445 | 5 | |
| Deletion | Chr9 | 22738001 | 22794000 | 56000 | 13 |
| Deletion | Chr1 | 23105445 | 23105456 | 11 | LOC_Os01g40860 |
| Deletion | Chr12 | 24207515 | 24207519 | 4 | |
| Deletion | Chr12 | 24345001 | 24399000 | 54000 | 6 |
| Deletion | Chr6 | 24732189 | 24732190 | 1 | |
| Deletion | Chr2 | 28491816 | 28491847 | 31 | |
| Deletion | Chr4 | 29562444 | 29562445 | 1 | |
| Deletion | Chr3 | 35032123 | 35032125 | 2 |
Insertions: 6
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 8386399 | 8433304 | LOC_Os08g14090 |
| Inversion | Chr1 | 9342267 | 9438197 | LOC_Os01g16450 |
| Inversion | Chr12 | 24344522 | 24506439 | LOC_Os12g39690 |
| Inversion | Chr12 | 24399015 | 24506441 | 2 |
| Inversion | Chr1 | 29464634 | 31772844 | |
| Inversion | Chr1 | 30528628 | 31772844 | LOC_Os01g53130 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 8432143 | Chr5 | 5868718 | LOC_Os08g14090 |
| Translocation | Chr8 | 8433273 | Chr5 | 5868715 | LOC_Os08g14090 |
| Translocation | Chr12 | 12485824 | Chr1 | 29464161 | |
| Translocation | Chr8 | 14295491 | Chr1 | 2949201 | LOC_Os08g23620 |
