Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN942-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN942-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 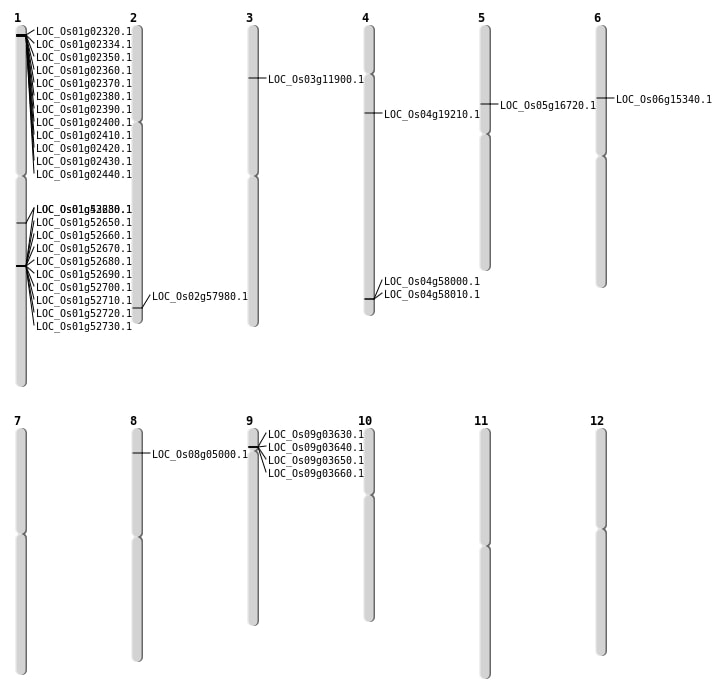
|
| Variant Information |
|---|
Single base substitutions: 23
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18892457 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 24739687 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g43280 |
| SBS | Chr1 | 37019426 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 3905569 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 13585047 | C-T | HOMO | INTRON | |
| SBS | Chr10 | 5810742 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 12641080 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 19901100 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 12860592 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 12860593 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 3133510 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 4505540 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 10901432 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 14795365 | A-G | HOMO | INTRON | |
| SBS | Chr3 | 28183380 | A-C | HET | INTRON | |
| SBS | Chr4 | 10674894 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g19210 |
| SBS | Chr4 | 16940965 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 9506932 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g16720 |
| SBS | Chr6 | 17859006 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 13736544 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 17207359 | T-C | HET | INTRON | |
| SBS | Chr7 | 9381750 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 1848647 | C-G | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 734001 | 792000 | 58000 | 12 |
| Deletion | Chr9 | 1816001 | 1844000 | 28000 | 4 |
| Deletion | Chr4 | 2029095 | 2029110 | 15 | |
| Deletion | Chr7 | 2125728 | 2125738 | 10 | |
| Deletion | Chr11 | 4005225 | 4005249 | 24 | |
| Deletion | Chr3 | 6233244 | 6233248 | 4 | LOC_Os03g11900 |
| Deletion | Chr12 | 7515177 | 7515183 | 6 | |
| Deletion | Chr7 | 7667136 | 7667137 | 1 | |
| Deletion | Chr6 | 7719574 | 7719576 | 2 | |
| Deletion | Chr8 | 8130054 | 8130056 | 2 | |
| Deletion | Chr1 | 10398956 | 10398963 | 7 | |
| Deletion | Chr1 | 11927990 | 11927994 | 4 | |
| Deletion | Chr6 | 24738679 | 24738681 | 2 | |
| Deletion | Chr7 | 26131557 | 26131562 | 5 | |
| Deletion | Chr8 | 26440550 | 26440552 | 2 | |
| Deletion | Chr2 | 29476416 | 29476417 | 1 | |
| Deletion | Chr1 | 30254001 | 30334000 | 80000 | 11 |
| Deletion | Chr4 | 34540182 | 34546462 | 6280 | 2 |
| Deletion | Chr2 | 34712362 | 34712368 | 6 | |
| Deletion | Chr2 | 35505950 | 35505951 | 1 | LOC_Os02g57980 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 1506840 | 10328813 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2564206 | Chr6 | 8708932 | 2 |
| Translocation | Chr8 | 2566502 | Chr6 | 8708928 | LOC_Os06g15340 |
| Translocation | Chr7 | 19449228 | Chr1 | 37074601 | |
| Translocation | Chr6 | 26040334 | Chr5 | 8931136 | |
| Translocation | Chr6 | 26040448 | Chr5 | 8931130 |
