Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN982-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN982-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 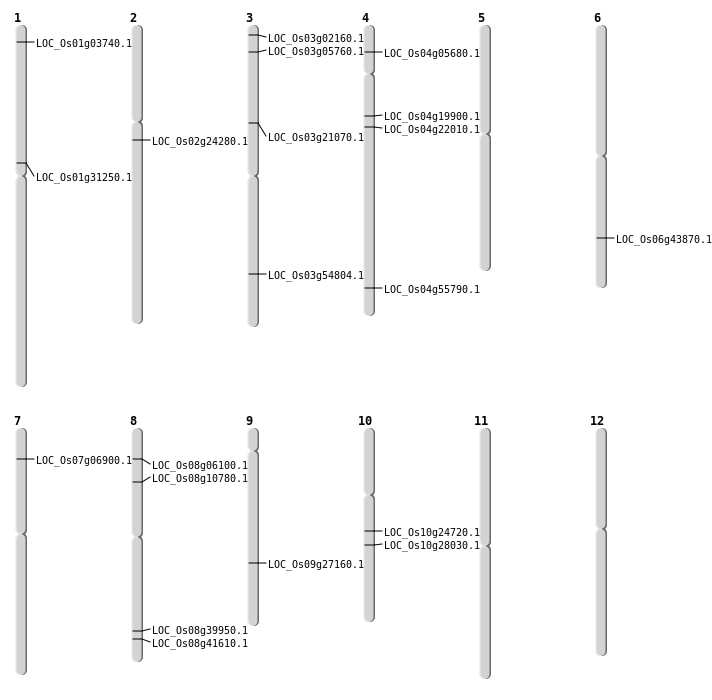
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1639507 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 17092285 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g31250 |
| SBS | Chr1 | 19748194 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 27192841 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 36395870 | C-G | HOMO | UTR_5_PRIME | |
| SBS | Chr1 | 36414279 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 12714023 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24720 |
| SBS | Chr10 | 14546323 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g28030 |
| SBS | Chr10 | 7438335 | C-A | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 15837213 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 22424130 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 292235 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 3697022 | A-T | HET | INTRON | |
| SBS | Chr3 | 10396214 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 11987576 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g21070 |
| SBS | Chr3 | 15491938 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 16325271 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 2660513 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 719845 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g02160 |
| SBS | Chr3 | 8173674 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 11097246 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g19900 |
| SBS | Chr4 | 12464231 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22010 |
| SBS | Chr4 | 17676636 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 2903469 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05680 |
| SBS | Chr4 | 29586993 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 7521408 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 4250240 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr5 | 9914183 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 24747460 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 18797515 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 27350841 | G-A | HET | INTRON | |
| SBS | Chr7 | 29205672 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 22421665 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 24891562 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr8 | 26277771 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g41610 |
| SBS | Chr8 | 6040193 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 18308614 | A-G | HET | INTRON | |
| SBS | Chr9 | 7767262 | T-G | HOMO | INTERGENIC |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 29340 | 29341 | 1 | |
| Deletion | Chr8 | 757574 | 757585 | 11 | |
| Deletion | Chr1 | 1564742 | 1564744 | 2 | LOC_Os01g03740 |
| Deletion | Chr7 | 3374135 | 3374136 | 1 | LOC_Os07g06900 |
| Deletion | Chr12 | 3696975 | 3696989 | 14 | |
| Deletion | Chr9 | 3739095 | 3739104 | 9 | |
| Deletion | Chr5 | 6160878 | 6160882 | 4 | |
| Deletion | Chr8 | 6339553 | 6339580 | 27 | LOC_Os08g10780 |
| Deletion | Chr10 | 7633688 | 7633701 | 13 | |
| Deletion | Chr10 | 9827590 | 9827700 | 110 | |
| Deletion | Chr7 | 11145734 | 11145735 | 1 | |
| Deletion | Chr10 | 13174971 | 13174976 | 5 | |
| Deletion | Chr2 | 14083025 | 14083030 | 5 | LOC_Os02g24280 |
| Deletion | Chr12 | 14157226 | 14157239 | 13 | |
| Deletion | Chr1 | 16314073 | 16314439 | 366 | |
| Deletion | Chr9 | 16516142 | 16516149 | 7 | LOC_Os09g27160 |
| Deletion | Chr4 | 16819263 | 16819264 | 1 | |
| Deletion | Chr2 | 18623280 | 18623285 | 5 | |
| Deletion | Chr11 | 18810310 | 18810311 | 1 | |
| Deletion | Chr3 | 19100379 | 19100391 | 12 | |
| Deletion | Chr8 | 20354493 | 20354497 | 4 | |
| Deletion | Chr12 | 22424109 | 22424153 | 44 | |
| Deletion | Chr8 | 25318128 | 25318135 | 7 | LOC_Os08g39950 |
| Deletion | Chr6 | 26421889 | 26421891 | 2 | LOC_Os06g43870 |
| Deletion | Chr3 | 26912341 | 26912342 | 1 | |
| Deletion | Chr6 | 28505141 | 28505142 | 1 | |
| Deletion | Chr4 | 30469606 | 30469608 | 2 | |
| Deletion | Chr3 | 31158543 | 31158552 | 9 | LOC_Os03g54804 |
| Deletion | Chr2 | 33203768 | 33203770 | 2 | |
| Deletion | Chr4 | 33214563 | 33214569 | 6 | LOC_Os04g55790 |
| Deletion | Chr1 | 37159245 | 37159249 | 4 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 6004294 | 6004294 | 1 | |
| Insertion | Chr3 | 24760679 | 24760680 | 2 | |
| Insertion | Chr8 | 11287759 | 11287759 | 1 | |
| Insertion | Chr8 | 26060431 | 26060431 | 1 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 3340280 | Chr3 | 2873109 | 2 |
| Translocation | Chr8 | 3342397 | Chr3 | 2873107 | LOC_Os03g05760 |
