Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN995-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN995-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 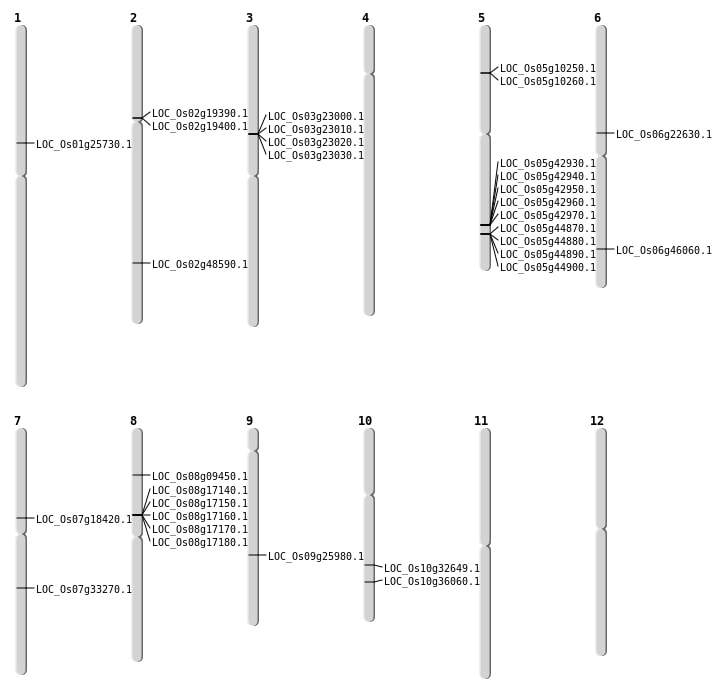
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13881301 | T-C | HET | INTRON | |
| SBS | Chr1 | 21530464 | A-G | HET | INTRON | |
| SBS | Chr1 | 21530465 | T-A | HET | INTRON | |
| SBS | Chr1 | 34497616 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 37457063 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 17105322 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g32649 |
| SBS | Chr10 | 19280804 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g36060 |
| SBS | Chr10 | 19350880 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 236602 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 19603350 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 28336505 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 16348732 | C-T | HET | INTRON | |
| SBS | Chr3 | 19534251 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 2656107 | C-T | HET | INTRON | |
| SBS | Chr3 | 28265334 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 36316031 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 4595530 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 4595531 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 24151757 | A-T | HET | INTRON | |
| SBS | Chr4 | 5433007 | T-A | HET | INTRON | |
| SBS | Chr5 | 22768029 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 9549230 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 13157022 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13157023 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g22630 |
| SBS | Chr7 | 10902128 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g18420 |
| SBS | Chr8 | 21369684 | C-T | HET | INTRON | |
| SBS | Chr8 | 5477280 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g09450 |
| SBS | Chr9 | 15819792 | C-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 17376632 | C-T | HOMO | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 2606339 | 2606340 | 1 | |
| Deletion | Chr5 | 5572001 | 5584000 | 12000 | 2 |
| Deletion | Chr7 | 7955006 | 7955007 | 1 | |
| Deletion | Chr4 | 8682111 | 8682112 | 1 | |
| Deletion | Chr8 | 10491501 | 10534302 | 42801 | 5 |
| Deletion | Chr3 | 10844052 | 10844053 | 1 | |
| Deletion | Chr2 | 11326001 | 11335000 | 9000 | 2 |
| Deletion | Chr6 | 13024891 | 13024899 | 8 | |
| Deletion | Chr3 | 13276001 | 13298000 | 22000 | 4 |
| Deletion | Chr8 | 13706635 | 13706636 | 1 | |
| Deletion | Chr1 | 14583116 | 14583127 | 11 | LOC_Os01g25730 |
| Deletion | Chr11 | 15296372 | 15296373 | 1 | |
| Deletion | Chr7 | 17180970 | 17180976 | 6 | |
| Deletion | Chr4 | 17563377 | 17563384 | 7 | |
| Deletion | Chr9 | 19349268 | 19349275 | 7 | |
| Deletion | Chr3 | 20910591 | 20910594 | 3 | |
| Deletion | Chr10 | 21591680 | 21591695 | 15 | |
| Deletion | Chr8 | 22700990 | 22700991 | 1 | |
| Deletion | Chr1 | 24765629 | 24765633 | 4 | |
| Deletion | Chr5 | 24932001 | 24955000 | 23000 | 5 |
| Deletion | Chr7 | 24992952 | 24992953 | 1 | |
| Deletion | Chr5 | 26072001 | 26095000 | 23000 | 4 |
| Deletion | Chr6 | 27924993 | 27924999 | 6 | LOC_Os06g46060 |
| Deletion | Chr2 | 30597335 | 30605231 | 7896 | |
| Deletion | Chr3 | 31027010 | 31027011 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 7567958 | 7567958 | 1 | |
| Insertion | Chr3 | 14962291 | 14962292 | 2 | |
| Insertion | Chr4 | 33654286 | 33654286 | 1 | |
| Insertion | Chr9 | 15606405 | 15606405 | 1 | LOC_Os09g25980 |
No Inversion
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1324219 | Chr2 | 29746299 | LOC_Os02g48590 |
| Translocation | Chr9 | 1330290 | Chr2 | 29742887 | |
| Translocation | Chr7 | 19888022 | Chr2 | 30604156 | LOC_Os07g33270 |
| Translocation | Chr7 | 19888060 | Chr2 | 30605256 | LOC_Os07g33270 |
