Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | W42-1-3S-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download W42-1-3S-1 Alignment File |
| Seed Availability | No |
| Mapping (Hover to Zoom-In) | 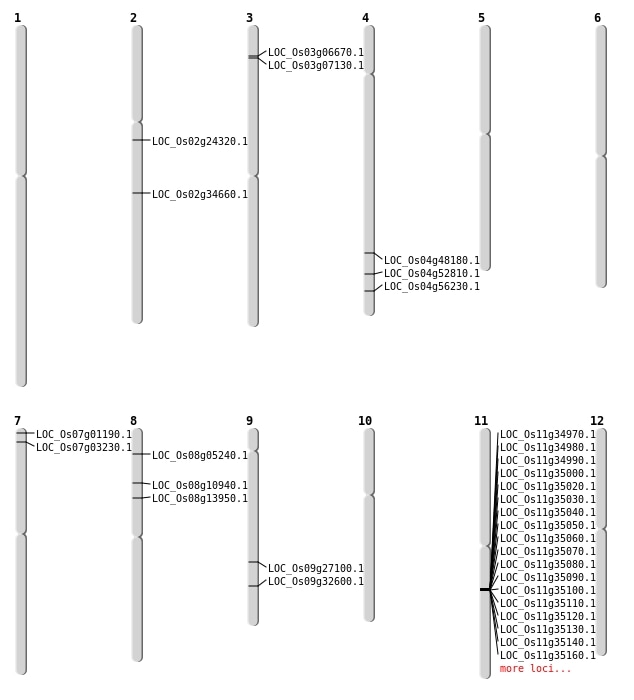
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12972454 | A-G | HET | INTRON | |
| SBS | Chr1 | 22969444 | T-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 30498492 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 11615058 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 12606323 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 137555 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 15694273 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 15954531 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 25251153 | A-C | HOMO | INTRON | |
| SBS | Chr11 | 25277760 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g42030 |
| SBS | Chr2 | 14110163 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g24320 |
| SBS | Chr2 | 14226688 | A-G | HET | INTRON | |
| SBS | Chr2 | 20792319 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g34660 |
| SBS | Chr2 | 2082542 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 30360195 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 33989338 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 6418512 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 3819347 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 3819350 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 13317262 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 13618613 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 13888214 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 17251170 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 19800939 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 25966891 | T-G | HET | INTRON | |
| SBS | Chr4 | 33525216 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g56230 |
| SBS | Chr4 | 33876539 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 6542135 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 1965199 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 19989421 | T-A | HOMO | INTRON | |
| SBS | Chr5 | 26229666 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 26955732 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr6 | 10121500 | T-G | HET | INTRON | |
| SBS | Chr6 | 7366681 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 114251 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g01190 |
| SBS | Chr7 | 1278230 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g03230 |
| SBS | Chr7 | 4388109 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 9410592 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 14680037 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 22384296 | G-C | HET | INTRON | |
| SBS | Chr8 | 3005370 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 8359394 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g13950 |
| SBS | Chr9 | 16441924 | G-A | HET | INTRON |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 72524 | 72525 | 1 | |
| Deletion | Chr4 | 98826 | 98842 | 16 | |
| Deletion | Chr3 | 213826 | 213829 | 3 | |
| Deletion | Chr3 | 3647018 | 3647026 | 8 | LOC_Os03g07130 |
| Deletion | Chr8 | 6453925 | 6453929 | 4 | LOC_Os08g10940 |
| Deletion | Chr8 | 6523397 | 6523398 | 1 | |
| Deletion | Chr10 | 7622296 | 7622308 | 12 | |
| Deletion | Chr4 | 10264003 | 10264012 | 9 | |
| Deletion | Chr12 | 11341748 | 11341749 | 1 | |
| Deletion | Chr7 | 13302011 | 13302012 | 1 | |
| Deletion | Chr7 | 13720364 | 13720367 | 3 | |
| Deletion | Chr1 | 18839489 | 18839499 | 10 | |
| Deletion | Chr7 | 20360528 | 20360545 | 17 | |
| Deletion | Chr11 | 20499001 | 20626000 | 127000 | 18 |
| Deletion | Chr11 | 20633001 | 20663000 | 30000 | 6 |
| Deletion | Chr11 | 20667001 | 20864000 | 197000 | 33 |
| Deletion | Chr4 | 21659274 | 21659288 | 14 | |
| Deletion | Chr1 | 22969672 | 22969673 | 1 | |
| Deletion | Chr1 | 26327989 | 26327991 | 2 | |
| Deletion | Chr3 | 27094736 | 27094738 | 2 | |
| Deletion | Chr1 | 28634340 | 28634775 | 435 | |
| Deletion | Chr4 | 28679863 | 28693250 | 13387 | LOC_Os04g48180 |
| Deletion | Chr4 | 33876542 | 33876549 | 7 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 15152328 | 15152328 | 1 | |
| Insertion | Chr11 | 18493101 | 18493101 | 1 | |
| Insertion | Chr2 | 34603953 | 34603953 | 1 | |
| Insertion | Chr4 | 31443242 | 31443256 | 15 | LOC_Os04g52810 |
| Insertion | Chr6 | 22127887 | 22127888 | 2 | |
| Insertion | Chr9 | 22500019 | 22500019 | 1 | |
| Insertion | Chr9 | 9495466 | 9495467 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 3371807 | 3977479 | LOC_Os03g06670 |
| Inversion | Chr9 | 16353318 | 16441923 | LOC_Os09g27030 |
| Inversion | Chr9 | 16353360 | 16488856 | LOC_Os09g27100 |
| Inversion | Chr11 | 19517658 | 20499240 | LOC_Os11g34970 |
| Inversion | Chr11 | 20811535 | 20865122 | LOC_Os11g35510 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 8864140 | Chr4 | 28679874 | |
| Translocation | Chr5 | 8864141 | Chr1 | 28634768 | |
| Translocation | Chr9 | 16488835 | Chr8 | 2726760 | 2 |
| Translocation | Chr9 | 19456620 | Chr8 | 2727712 | 2 |
| Translocation | Chr4 | 28684629 | Chr1 | 28634350 |
