Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | CW-631-3-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download CW-631-3-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 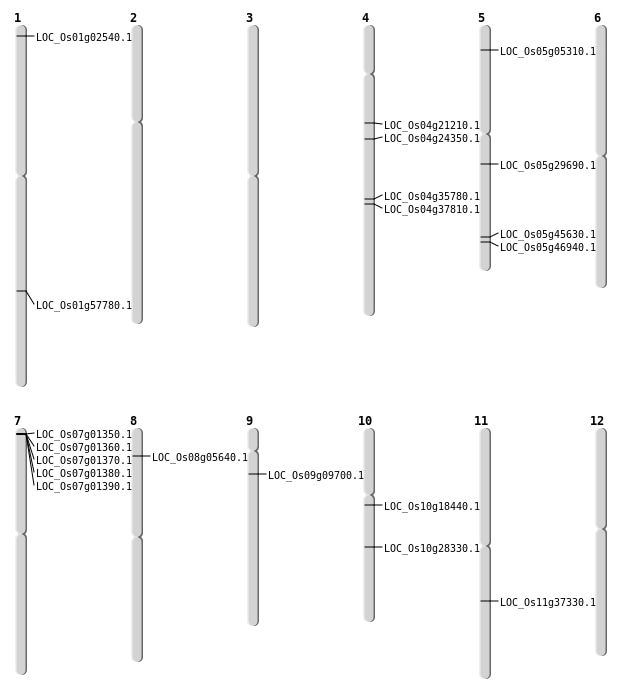
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 35563753 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 835024 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g02540 |
| SBS | Chr10 | 10126978 | A-G | HET | INTRON | |
| SBS | Chr11 | 4616114 | A-G | HET | INTRON | |
| SBS | Chr12 | 21099614 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 2293011 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 26244838 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 4266649 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 15706974 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 15706975 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 28211932 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 28309002 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 17648050 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 18995145 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 21479777 | A-G | HET | INTRON | |
| SBS | Chr4 | 11957279 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g21210 |
| SBS | Chr4 | 13953705 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g24350 |
| SBS | Chr4 | 16253117 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 21799699 | A-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g35780 |
| SBS | Chr4 | 22472393 | G-A | HOMO | SPLICE_SITE_DONOR | LOC_Os04g37810 |
| SBS | Chr4 | 25671480 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 7422988 | A-G | HET | INTRON | |
| SBS | Chr5 | 17005141 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 17161563 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g29690 |
| SBS | Chr5 | 17713036 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20566450 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20566453 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 21345576 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 26447541 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g45630 |
| SBS | Chr5 | 27162788 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g46940 |
| SBS | Chr5 | 2805418 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 5273020 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 121978 | G-A | HET | INTRON | |
| SBS | Chr6 | 14114426 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 23186122 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 26383494 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 4099960 | T-C | HET | INTRON | |
| SBS | Chr7 | 22685770 | C-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 24005781 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 9947197 | G-C | HET | INTRON | |
| SBS | Chr8 | 1121312 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr8 | 15509767 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 3015480 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g05640 |
| SBS | Chr9 | 5249895 | C-T | HOMO | SYNONYMOUS_CODING |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 222140 | 252470 | 30330 | 5 |
| Deletion | Chr10 | 2384055 | 2384071 | 16 | |
| Deletion | Chr5 | 2617229 | 2617231 | 2 | LOC_Os05g05310 |
| Deletion | Chr1 | 7694893 | 7694896 | 3 | |
| Deletion | Chr10 | 9372033 | 9372040 | 7 | LOC_Os10g18440 |
| Deletion | Chr11 | 19997450 | 19997453 | 3 | |
| Deletion | Chr8 | 21225957 | 21225961 | 4 | |
| Deletion | Chr10 | 21972148 | 21972156 | 8 | |
| Deletion | Chr11 | 22043088 | 22043089 | 1 | LOC_Os11g37330 |
| Deletion | Chr8 | 23859495 | 23859497 | 2 | |
| Deletion | Chr1 | 27027854 | 27027856 | 2 | |
| Deletion | Chr1 | 27073981 | 27073983 | 2 | |
| Deletion | Chr5 | 27532070 | 27532071 | 1 | |
| Deletion | Chr7 | 28242704 | 28242709 | 5 | |
| Deletion | Chr1 | 33418980 | 33418986 | 6 | LOC_Os01g57780 |
| Deletion | Chr1 | 35982104 | 35982555 | 451 |
Insertions: 5
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 4830074 | 5247232 | LOC_Os09g09700 |
| Inversion | Chr9 | 5247231 | 7081975 | LOC_Os09g09700 |
| Inversion | Chr10 | 11646694 | 14742428 | LOC_Os10g28330 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 10776825 | Chr4 | 20488252 | |
| Translocation | Chr6 | 10777922 | Chr4 | 20488273 | |
| Translocation | Chr8 | 11601768 | Chr4 | 61838 | |
| Translocation | Chr4 | 18850944 | Chr1 | 35982536 | |
| Translocation | Chr4 | 18850961 | Chr1 | 35982110 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr2 | 34291247 | 34291555 | 308 |
