Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-100 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-100 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 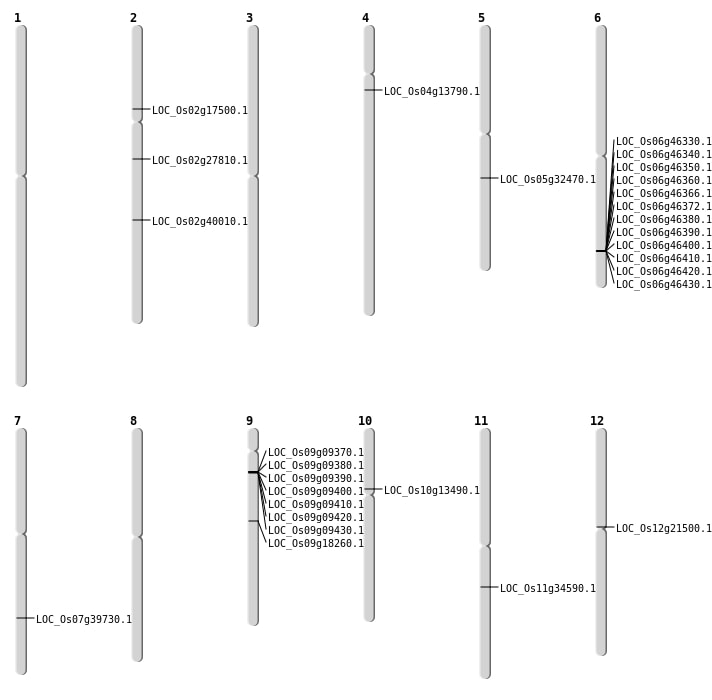
|
| Variant Information |
|---|
Single base substitutions: 22
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26457437 | C-G | HET | INTRON | |
| SBS | Chr1 | 33603139 | A-G | HOMO | INTRON | |
| SBS | Chr10 | 7290965 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g13490 |
| SBS | Chr11 | 22267045 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 21099208 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 3751589 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 12171967 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 15581488 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 7338024 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 20833084 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 7694364 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13790 |
| SBS | Chr5 | 26632967 | G-A | HET | INTRON | |
| SBS | Chr5 | 26632968 | G-A | HET | INTRON | |
| SBS | Chr6 | 28032272 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 23812120 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g39730 |
| SBS | Chr8 | 12143256 | T-A | HET | INTRON | |
| SBS | Chr9 | 11213067 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g18260 |
| SBS | Chr9 | 16548385 | A-G | HET | INTRON | |
| SBS | Chr9 | 16649712 | T-G | HET | INTRON | |
| SBS | Chr9 | 3462914 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 9950753 | G-A | HOMO | INTRON | |
| SBS | Chr9 | 9950754 | G-A | HOMO | INTRON |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 206390 | 206392 | 2 | |
| Deletion | Chr9 | 551138 | 551141 | 3 | |
| Deletion | Chr9 | 2900675 | 2900691 | 16 | |
| Deletion | Chr9 | 5023001 | 5083000 | 60000 | 7 |
| Deletion | Chr5 | 6699683 | 6699691 | 8 | |
| Deletion | Chr1 | 6796203 | 6796205 | 2 | |
| Deletion | Chr1 | 9177630 | 9177632 | 2 | |
| Deletion | Chr9 | 9222495 | 9222496 | 1 | |
| Deletion | Chr1 | 9906310 | 9906314 | 4 | |
| Deletion | Chr2 | 10074733 | 10074739 | 6 | LOC_Os02g17500 |
| Deletion | Chr8 | 11308989 | 11308992 | 3 | |
| Deletion | Chr8 | 11473363 | 11473364 | 1 | |
| Deletion | Chr12 | 12079394 | 12079402 | 8 | LOC_Os12g21500 |
| Deletion | Chr11 | 14309180 | 14309187 | 7 | |
| Deletion | Chr9 | 15024634 | 15024639 | 5 | |
| Deletion | Chr7 | 16416143 | 16416146 | 3 | |
| Deletion | Chr5 | 18965497 | 18965533 | 36 | LOC_Os05g32470 |
| Deletion | Chr4 | 23983124 | 23983131 | 7 | |
| Deletion | Chr11 | 24570374 | 24570391 | 17 | |
| Deletion | Chr4 | 26088878 | 26088879 | 1 | |
| Deletion | Chr12 | 27040159 | 27040160 | 1 | |
| Deletion | Chr6 | 28084001 | 28180000 | 96000 | 12 |
| Deletion | Chr5 | 28136091 | 28136103 | 12 | |
| Deletion | Chr4 | 30425712 | 30425718 | 6 |
Insertions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11545605 | 11545606 | 2 | |
| Insertion | Chr1 | 23308315 | 23308320 | 6 | |
| Insertion | Chr1 | 23572413 | 23572414 | 2 | |
| Insertion | Chr1 | 26649035 | 26649035 | 1 | |
| Insertion | Chr1 | 35237639 | 35237646 | 8 | |
| Insertion | Chr1 | 40468997 | 40468999 | 3 | |
| Insertion | Chr11 | 14361450 | 14361450 | 1 | |
| Insertion | Chr11 | 20246983 | 20246983 | 1 | LOC_Os11g34590 |
| Insertion | Chr11 | 27699077 | 27699078 | 2 | |
| Insertion | Chr12 | 4445173 | 4445173 | 1 | |
| Insertion | Chr12 | 8085373 | 8085376 | 4 | |
| Insertion | Chr2 | 16468454 | 16468457 | 4 | LOC_Os02g27810 |
| Insertion | Chr2 | 16479231 | 16479242 | 12 | |
| Insertion | Chr2 | 18552323 | 18552324 | 2 | |
| Insertion | Chr2 | 24228585 | 24228590 | 6 | LOC_Os02g40010 |
| Insertion | Chr2 | 29896930 | 29896937 | 8 | |
| Insertion | Chr2 | 4064087 | 4064088 | 2 | |
| Insertion | Chr3 | 30631961 | 30631961 | 1 | |
| Insertion | Chr4 | 33475327 | 33475340 | 14 | |
| Insertion | Chr4 | 34230146 | 34230147 | 2 | |
| Insertion | Chr5 | 5291967 | 5291967 | 1 | |
| Insertion | Chr6 | 22279922 | 22279923 | 2 | |
| Insertion | Chr6 | 24037839 | 24037842 | 4 | |
| Insertion | Chr6 | 29472167 | 29472168 | 2 | |
| Insertion | Chr7 | 23386232 | 23386233 | 2 | |
| Insertion | Chr8 | 16099831 | 16099832 | 2 | |
| Insertion | Chr8 | 20070781 | 20070782 | 2 | |
| Insertion | Chr9 | 15169579 | 15169580 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 4667790 | 7267334 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 27136537 | Chr3 | 8253979 |
