Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-101 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-101 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 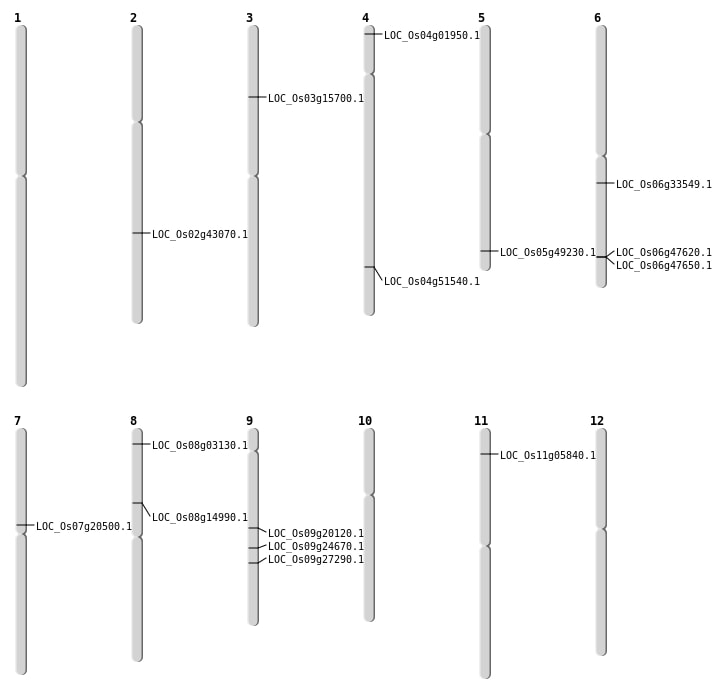
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 10298295 | A-T | HET | INTRON | |
| SBS | Chr10 | 4711945 | C-T | HET | INTRON | |
| SBS | Chr10 | 7292388 | C-T | HET | INTRON | |
| SBS | Chr10 | 8771288 | G-A | HET | INTRON | |
| SBS | Chr11 | 20142735 | C-G | HET | INTRON | |
| SBS | Chr11 | 2723397 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g05840 |
| SBS | Chr11 | 7467996 | A-C | HET | INTRON | |
| SBS | Chr12 | 13580366 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 24539957 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 1007746 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 1007747 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 35857903 | A-T | HET | INTRON | |
| SBS | Chr2 | 3817750 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 18124810 | G-T | HET | INTRON | |
| SBS | Chr3 | 24182061 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 29943847 | G-A | HET | INTRON | |
| SBS | Chr3 | 8660589 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g15700 |
| SBS | Chr4 | 13449686 | G-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 24004 | T-G | HOMO | INTRON | |
| SBS | Chr4 | 30535883 | T-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 731213 | A-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 11525573 | G-T | HET | INTRON | |
| SBS | Chr5 | 28248991 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g49230 |
| SBS | Chr6 | 14414826 | A-G | HET | INTRON | |
| SBS | Chr6 | 25598911 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 11838503 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g20500 |
| SBS | Chr8 | 10397201 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 1433231 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g03130 |
| SBS | Chr8 | 14975239 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 12058290 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g20120 |
| SBS | Chr9 | 14683352 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g24670 |
| SBS | Chr9 | 14683353 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 1642574 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 16592052 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g27290 |
| SBS | Chr9 | 8339132 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 9225941 | G-A | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 594354 | 594358 | 4 | LOC_Os04g01950 |
| Deletion | Chr2 | 1054834 | 1054835 | 1 | |
| Deletion | Chr9 | 5576772 | 5576777 | 5 | |
| Deletion | Chr8 | 9043107 | 9043123 | 16 | LOC_Os08g14990 |
| Deletion | Chr4 | 9650840 | 9650841 | 1 | |
| Deletion | Chr1 | 10353581 | 10353582 | 1 | |
| Deletion | Chr5 | 10640184 | 10640185 | 1 | |
| Deletion | Chr11 | 11023347 | 11023363 | 16 | |
| Deletion | Chr9 | 14150143 | 14150144 | 1 | |
| Deletion | Chr9 | 14526040 | 14526046 | 6 | |
| Deletion | Chr11 | 14995103 | 14995114 | 11 | |
| Deletion | Chr2 | 15189151 | 15189157 | 6 | |
| Deletion | Chr7 | 15882754 | 15882755 | 1 | |
| Deletion | Chr6 | 17381786 | 17381787 | 1 | |
| Deletion | Chr6 | 18071108 | 18071127 | 19 | |
| Deletion | Chr12 | 18359156 | 18359158 | 2 | |
| Deletion | Chr7 | 20785138 | 20785149 | 11 | |
| Deletion | Chr10 | 21072176 | 21072183 | 7 | |
| Deletion | Chr7 | 22207950 | 22207952 | 2 | |
| Deletion | Chr2 | 25937298 | 25937309 | 11 | LOC_Os02g43070 |
| Deletion | Chr4 | 30535984 | 30535985 | 1 | LOC_Os04g51540 |
| Deletion | Chr2 | 30686697 | 30686698 | 1 | |
| Deletion | Chr3 | 35328956 | 35329048 | 92 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr7 | 14740466 | 14740466 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 10763445 | 12844188 | |
| Inversion | Chr10 | 12844176 | 20818783 | |
| Inversion | Chr6 | 18205986 | 19541549 | LOC_Os06g33549 |
| Inversion | Chr6 | 28834746 | 28856181 | 2 |
| Inversion | Chr6 | 28856182 | 29493270 | LOC_Os06g47650 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 23188874 | Chr6 | 29493264 | |
| Translocation | Chr11 | 23188890 | Chr6 | 28834784 | LOC_Os06g47620 |
