Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-13 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-13 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 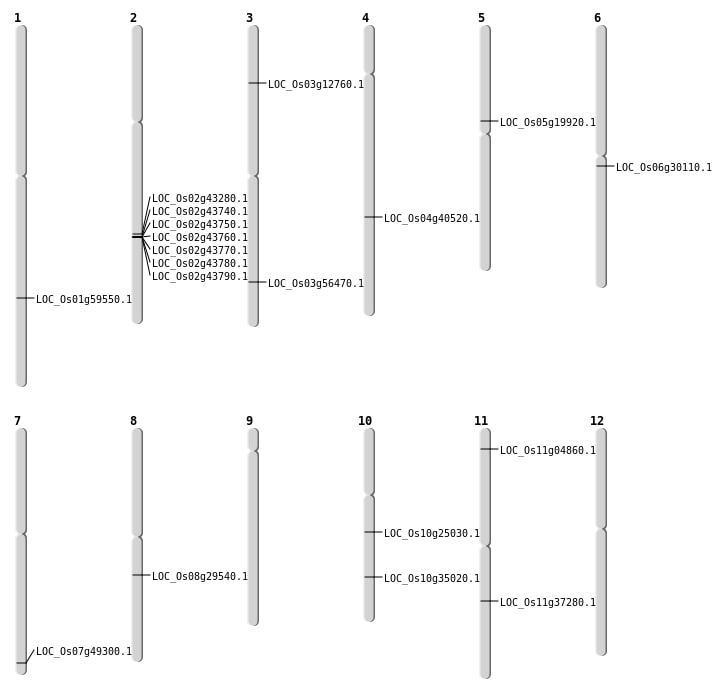
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 34432688 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g59550 |
| SBS | Chr11 | 16214207 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 16214207 | G-A | Homo | INTERGENIC | |
| SBS | Chr12 | 21433094 | C-T | HET | INTRON | |
| SBS | Chr12 | 4229865 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 8181001 | A-G | HET | INTRON | |
| SBS | Chr3 | 32183255 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g56470 |
| SBS | Chr3 | 32183255 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g56470 |
| SBS | Chr3 | 32183256 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 32183256 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr3 | 4216246 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 4216246 | T-C | Homo | INTERGENIC | |
| SBS | Chr4 | 1428248 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 1428248 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 26553470 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 26553470 | G-A | Homo | INTERGENIC | |
| SBS | Chr5 | 27006277 | C-G | HOMO | INTRON | |
| SBS | Chr5 | 27006277 | C-G | Homo | INTRON | |
| SBS | Chr5 | 28117991 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 28227595 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr6 | 27600249 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 27600249 | T-C | Homo | INTERGENIC | |
| SBS | Chr6 | 7905511 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 6889642 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 7537271 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 7605918 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 10660800 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 695114 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 2223683 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 3524223 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 3524227 | C-A | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 498724 | 498725 | 1 | |
| Deletion | Chr11 | 2068509 | 2068566 | 57 | LOC_Os11g04860 |
| Deletion | Chr10 | 3478370 | 3478371 | 1 | |
| Deletion | Chr8 | 3664217 | 3664258 | 41 | |
| Deletion | Chr9 | 4741880 | 4741882 | 2 | |
| Deletion | Chr8 | 4975866 | 4975875 | 9 | |
| Deletion | Chr3 | 5180534 | 5180535 | 1 | |
| Deletion | Chr8 | 9390835 | 9390879 | 44 | |
| Deletion | Chr5 | 11620670 | 11620678 | 8 | LOC_Os05g19920 |
| Deletion | Chr4 | 11802533 | 11802537 | 4 | |
| Deletion | Chr11 | 13091414 | 13091415 | 1 | |
| Deletion | Chr6 | 17357965 | 17358391 | 426 | LOC_Os06g30110 |
| Deletion | Chr2 | 17569743 | 17569753 | 10 | |
| Deletion | Chr10 | 18683254 | 18683259 | 5 | LOC_Os10g35020 |
| Deletion | Chr1 | 19336413 | 19336414 | 1 | |
| Deletion | Chr4 | 24070863 | 24070884 | 21 | LOC_Os04g40520 |
| Deletion | Chr2 | 26395001 | 26424000 | 29000 | 6 |
| Deletion | Chr1 | 27917120 | 27917131 | 11 | |
| Deletion | Chr7 | 29524416 | 29524417 | 1 | LOC_Os07g49300 |
| Deletion | Chr2 | 29602452 | 29602470 | 18 | |
| Deletion | Chr2 | 30357939 | 30357940 | 1 | |
| Deletion | Chr4 | 35102250 | 35102258 | 8 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr7 | 11697414 | 11697415 | 2 | |
| Insertion | Chr9 | 18860673 | 18860673 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 6828848 | 9036485 | LOC_Os03g12760 |
| Inversion | Chr11 | 22017719 | 22136667 | LOC_Os11g37280 |
| Inversion | Chr11 | 22017733 | 22136667 | LOC_Os11g37280 |
| Inversion | Chr2 | 26082213 | 26394424 | 2 |
| Inversion | Chr2 | 26082227 | 26424143 | LOC_Os02g43280 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 12883297 | Chr8 | 18114123 | LOC_Os08g29540 |
| Translocation | Chr10 | 12886575 | Chr8 | 18114124 | 2 |
| Translocation | Chr11 | 20024974 | Chr7 | 4983063 |
