Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-169 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-169 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 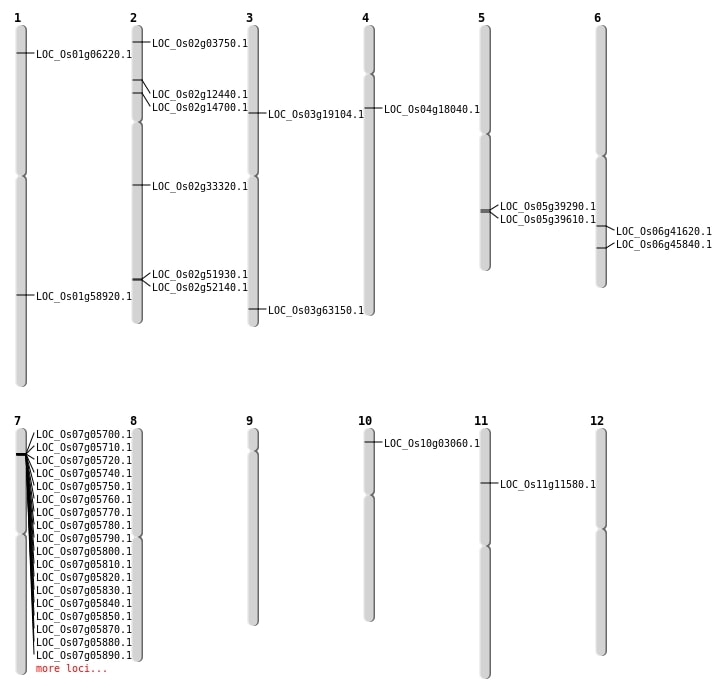
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10103281 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 1180527 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 15490895 | T-G | HET | INTRON | |
| SBS | Chr1 | 2957533 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g06220 |
| SBS | Chr1 | 29881343 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 34057664 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g58920 |
| SBS | Chr10 | 1167154 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 1276121 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g03060 |
| SBS | Chr10 | 17836969 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 20356637 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 20743565 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 3600495 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 7201237 | T-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 14940690 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 9637283 | T-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 10704768 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g19104 |
| SBS | Chr3 | 20299332 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 29498919 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 34478278 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr3 | 34984648 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 19614018 | C-T | HET | INTRON | |
| SBS | Chr4 | 26872573 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 32049011 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 5928583 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 23038580 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g39290 |
| SBS | Chr5 | 28282699 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 6137784 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 1906219 | A-G | HET | INTRON | |
| SBS | Chr6 | 20282442 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 24927257 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g41620 |
| SBS | Chr6 | 27735479 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g45840 |
| SBS | Chr6 | 4931223 | C-T | HET | INTRON | |
| SBS | Chr6 | 5496827 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 11252901 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2718496 | A-G | HET | INTRON | |
| SBS | Chr7 | 778740 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 15748136 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 19208050 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 20239438 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 28325641 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 13866239 | C-T | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1572795 | 1574353 | 1558 | LOC_Os02g03750 |
| Deletion | Chr7 | 2723001 | 2917000 | 194000 | 30 |
| Deletion | Chr10 | 3792308 | 3792309 | 1 | |
| Deletion | Chr11 | 6442030 | 6442074 | 44 | LOC_Os11g11580 |
| Deletion | Chr2 | 6490377 | 6490381 | 4 | LOC_Os02g12440 |
| Deletion | Chr2 | 8107875 | 8107885 | 10 | LOC_Os02g14700 |
| Deletion | Chr6 | 8192846 | 8192847 | 1 | |
| Deletion | Chr9 | 8915585 | 8915589 | 4 | |
| Deletion | Chr4 | 9967519 | 9967520 | 1 | LOC_Os04g18040 |
| Deletion | Chr11 | 10008456 | 10008463 | 7 | |
| Deletion | Chr2 | 10868372 | 10868373 | 1 | |
| Deletion | Chr6 | 11924182 | 11924765 | 583 | |
| Deletion | Chr7 | 13990001 | 14230000 | 240000 | 39 |
| Deletion | Chr7 | 14245001 | 14291000 | 46000 | 4 |
| Deletion | Chr5 | 14731482 | 14731486 | 4 | |
| Deletion | Chr2 | 19794020 | 19794021 | 1 | LOC_Os02g33320 |
| Deletion | Chr5 | 21242814 | 21242815 | 1 | |
| Deletion | Chr3 | 22081564 | 22081567 | 3 | |
| Deletion | Chr12 | 22243062 | 22243063 | 1 | |
| Deletion | Chr5 | 23255004 | 23255006 | 2 | LOC_Os05g39610 |
| Deletion | Chr3 | 23538166 | 23538177 | 11 | |
| Deletion | Chr7 | 24980457 | 24980458 | 1 | |
| Deletion | Chr4 | 33924314 | 33924324 | 10 | |
| Deletion | Chr3 | 35694799 | 35694800 | 1 | LOC_Os03g63150 |
| Deletion | Chr1 | 40916356 | 40916357 | 1 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 24063913 | 24183912 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 3373501 | Chr2 | 31920134 | 2 |
| Translocation | Chr7 | 3373534 | Chr2 | 31807763 | 2 |
| Translocation | Chr10 | 5530573 | Chr6 | 11924763 |
