Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-257 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-257 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 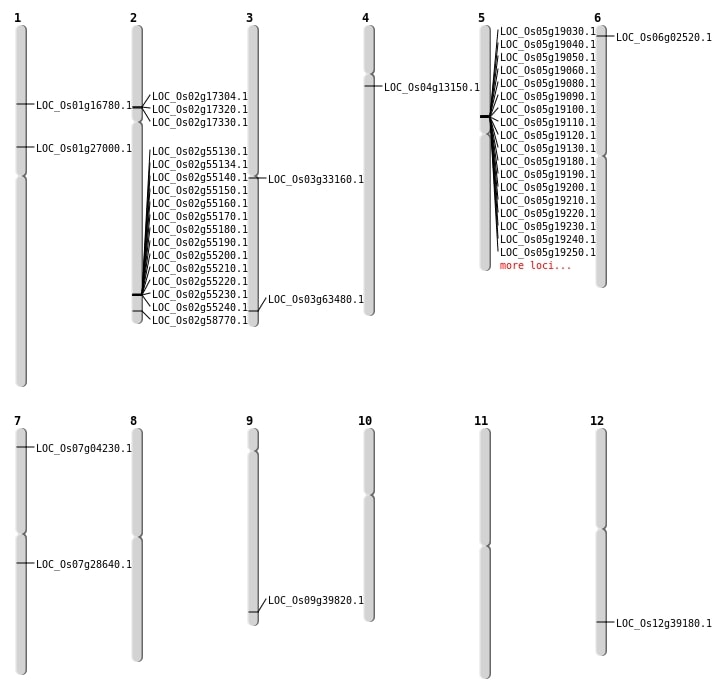
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15043653 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g27000 |
| SBS | Chr1 | 22257673 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 24500602 | G-A | HET | INTRON | |
| SBS | Chr1 | 32216903 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 9186796 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 9535255 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g16780 |
| SBS | Chr12 | 14163548 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 18848325 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 24773504 | T-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 26900489 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 16011672 | G-T | HET | INTRON | |
| SBS | Chr3 | 18966160 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g33160 |
| SBS | Chr3 | 19452706 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 35863362 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g63480 |
| SBS | Chr4 | 24150247 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 35460686 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 9221395 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 8342190 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 13737072 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 17931790 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 1899335 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 29353189 | T-G | HET | INTRON | |
| SBS | Chr7 | 17023233 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 1860528 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g04230 |
| SBS | Chr7 | 2869208 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 5060176 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 13627985 | T-C | HOMO | INTRON |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 873806 | 873818 | 12 | LOC_Os06g02520 |
| Deletion | Chr6 | 1032187 | 1032188 | 1 | |
| Deletion | Chr6 | 2402904 | 2402905 | 1 | |
| Deletion | Chr6 | 4306195 | 4306200 | 5 | |
| Deletion | Chr10 | 7008948 | 7008949 | 1 | |
| Deletion | Chr4 | 7255123 | 7255149 | 26 | LOC_Os04g13150 |
| Deletion | Chr9 | 7503858 | 7503859 | 1 | |
| Deletion | Chr5 | 8313745 | 8313746 | 1 | |
| Deletion | Chr3 | 8956119 | 8956121 | 2 | |
| Deletion | Chr2 | 9924093 | 9950870 | 26777 | 3 |
| Deletion | Chr2 | 10369151 | 10369158 | 7 | |
| Deletion | Chr5 | 11041001 | 11101000 | 60000 | 10 |
| Deletion | Chr5 | 11126001 | 11570000 | 444000 | 64 |
| Deletion | Chr5 | 11594001 | 12074000 | 480000 | 66 |
| Deletion | Chr7 | 13058860 | 13058861 | 1 | |
| Deletion | Chr7 | 16763969 | 16764480 | 511 | LOC_Os07g28640 |
| Deletion | Chr7 | 17539680 | 17542249 | 2569 | |
| Deletion | Chr10 | 18140683 | 18140703 | 20 | |
| Deletion | Chr10 | 20775984 | 20775996 | 12 | |
| Deletion | Chr5 | 22065234 | 22065267 | 33 | |
| Deletion | Chr7 | 22209303 | 22209308 | 5 | |
| Deletion | Chr4 | 22311314 | 22311321 | 7 | |
| Deletion | Chr9 | 22834486 | 22834487 | 1 | LOC_Os09g39820 |
| Deletion | Chr12 | 24120965 | 24120984 | 19 | LOC_Os12g39180 |
| Deletion | Chr2 | 33774001 | 33830000 | 56000 | 13 |
| Deletion | Chr2 | 35901995 | 35902000 | 5 | LOC_Os02g58770 |
Insertions: 7
No Inversion
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 11331700 | Chr1 | 4670915 | |
| Translocation | Chr7 | 16763981 | Chr5 | 11327969 | LOC_Os07g28640 |
| Translocation | Chr7 | 16764476 | Chr1 | 4670922 | LOC_Os07g28640 |
