Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-288 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-288 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 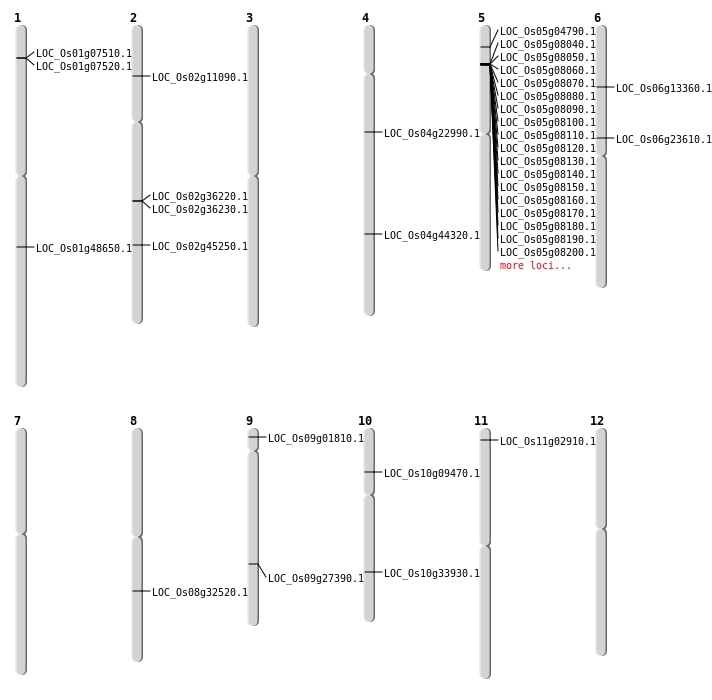
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21700157 | C-G | HOMO | INTRON | |
| SBS | Chr10 | 14939875 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1496875 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 1964329 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 5903637 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 6340935 | A-T | HET | INTRON | |
| SBS | Chr10 | 6340936 | A-T | HET | INTRON | |
| SBS | Chr10 | 6340938 | C-T | HET | INTRON | |
| SBS | Chr10 | 6347292 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 1414656 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 16040093 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 21168940 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 11337078 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 18201695 | G-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 19180588 | T-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 2596696 | C-T | HET | INTRON | |
| SBS | Chr12 | 3552594 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr12 | 8977313 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 21868229 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g36230 |
| SBS | Chr2 | 2791140 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 31077149 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 16781499 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 22256513 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 7385481 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 2277995 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g04790 |
| SBS | Chr6 | 11236786 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 9342439 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 13312423 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 20642112 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 23621992 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 26364505 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 3645799 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 4413697 | T-C | HET | INTRON | |
| SBS | Chr9 | 4125073 | C-T | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 560619 | 560621 | 2 | LOC_Os09g01810 |
| Deletion | Chr10 | 1454599 | 1454607 | 8 | |
| Deletion | Chr1 | 3583802 | 3595138 | 11336 | 2 |
| Deletion | Chr5 | 4347001 | 4525000 | 178000 | 26 |
| Deletion | Chr10 | 5108743 | 5108760 | 17 | LOC_Os10g09470 |
| Deletion | Chr8 | 7256437 | 7256441 | 4 | |
| Deletion | Chr6 | 7346237 | 7346240 | 3 | LOC_Os06g13360 |
| Deletion | Chr11 | 8836279 | 8836305 | 26 | |
| Deletion | Chr4 | 13051107 | 13051109 | 2 | LOC_Os04g22990 |
| Deletion | Chr6 | 13772387 | 13772388 | 1 | LOC_Os06g23610 |
| Deletion | Chr5 | 15991599 | 15991624 | 25 | |
| Deletion | Chr9 | 16646238 | 16646247 | 9 | LOC_Os09g27390 |
| Deletion | Chr1 | 17482111 | 17482112 | 1 | |
| Deletion | Chr10 | 18036051 | 18040061 | 4010 | LOC_Os10g33930 |
| Deletion | Chr2 | 18745749 | 18745750 | 1 | |
| Deletion | Chr7 | 19026030 | 19026032 | 2 | |
| Deletion | Chr8 | 22937211 | 22955391 | 18180 | |
| Deletion | Chr6 | 26298682 | 26298685 | 3 | |
| Deletion | Chr12 | 27333455 | 27333476 | 21 | |
| Deletion | Chr2 | 27488567 | 27488569 | 2 | LOC_Os02g45250 |
| Deletion | Chr2 | 27656410 | 27656417 | 7 | |
| Deletion | Chr4 | 27753065 | 27753066 | 1 | |
| Deletion | Chr1 | 27888068 | 27888072 | 4 | LOC_Os01g48650 |
| Deletion | Chr2 | 29819422 | 29819425 | 3 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12717709 | 12717709 | 1 | |
| Insertion | Chr11 | 7087730 | 7087731 | 2 | |
| Insertion | Chr2 | 12864063 | 12864066 | 4 | |
| Insertion | Chr3 | 459719 | 459719 | 1 | |
| Insertion | Chr4 | 26251100 | 26251100 | 1 | LOC_Os04g44320 |
| Insertion | Chr4 | 368582 | 368585 | 4 | |
| Insertion | Chr7 | 12108498 | 12108500 | 3 | |
| Insertion | Chr8 | 20145118 | 20145126 | 9 | LOC_Os08g32520 |
| Insertion | Chr8 | 26805875 | 26805878 | 4 | |
| Insertion | Chr9 | 8974005 | 8974006 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 6126479 | 18965196 | 2 |
| Inversion | Chr2 | 21698244 | 21860440 | LOC_Os02g36220 |
| Inversion | Chr2 | 21698267 | 21860530 | LOC_Os02g36220 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1002062 | Chr2 | 5944607 | 2 |
| Translocation | Chr7 | 9757780 | Chr4 | 3713103 | |
| Translocation | Chr4 | 11551033 | Chr2 | 17320461 | |
| Translocation | Chr12 | 25824844 | Chr1 | 1269276 | |
| Translocation | Chr12 | 25824852 | Chr1 | 1281057 |
