Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-289 [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-289 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 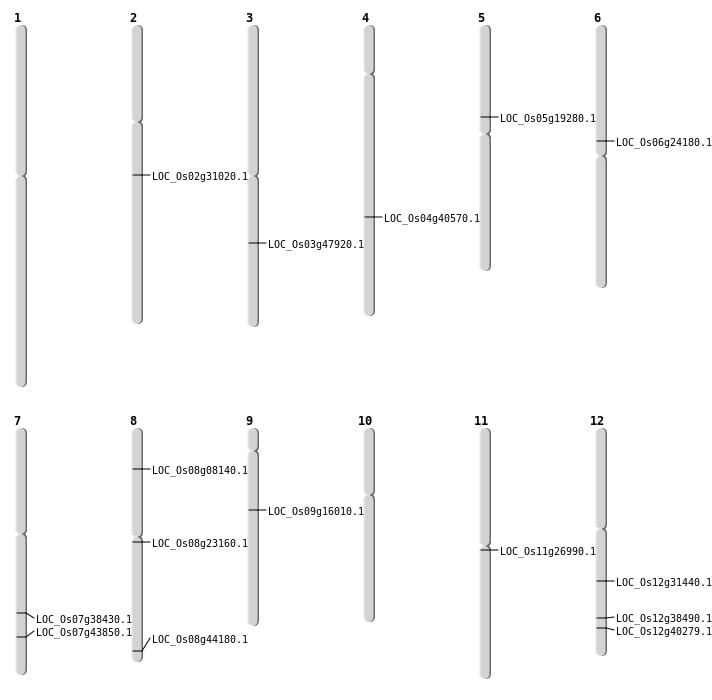
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16520981 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 37053060 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 37053063 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 37053064 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 15533177 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g26990 |
| SBS | Chr11 | 27298090 | G-A | HET | INTRON | |
| SBS | Chr11 | 8957979 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 18910896 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g31440 |
| SBS | Chr12 | 24947538 | A-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g40279 |
| SBS | Chr12 | 4986628 | T-A | HET | INTRON | |
| SBS | Chr12 | 591618 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 18545042 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 2086172 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 31414933 | T-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 3584575 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 7808337 | G-A | HET | INTRON | |
| SBS | Chr3 | 19696004 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 3288779 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 35194749 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 19632787 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 13208220 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 17276695 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 23092363 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g38430 |
| SBS | Chr7 | 26225044 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g43850 |
| SBS | Chr7 | 4171634 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 23619058 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 13756359 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 4180385 | T-A | HOMO | INTERGENIC |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1661711 | 1661716 | 5 | |
| Deletion | Chr12 | 3815590 | 3815598 | 8 | |
| Deletion | Chr8 | 4638304 | 4638316 | 12 | LOC_Os08g08140 |
| Deletion | Chr9 | 9783837 | 9783838 | 1 | LOC_Os09g16010 |
| Deletion | Chr1 | 13637565 | 13637567 | 2 | |
| Deletion | Chr8 | 17331287 | 17331291 | 4 | |
| Deletion | Chr8 | 18122360 | 18122362 | 2 | |
| Deletion | Chr2 | 18527923 | 18527952 | 29 | LOC_Os02g31020 |
| Deletion | Chr1 | 19606615 | 19606620 | 5 | |
| Deletion | Chr6 | 20875270 | 20875271 | 1 | |
| Deletion | Chr8 | 23146456 | 23146457 | 1 | |
| Deletion | Chr1 | 25173629 | 25173641 | 12 | |
| Deletion | Chr3 | 27534804 | 27534805 | 1 | |
| Deletion | Chr8 | 27814220 | 27814233 | 13 | LOC_Os08g44180 |
| Deletion | Chr1 | 28624713 | 28624714 | 1 | |
| Deletion | Chr5 | 28705233 | 28705254 | 21 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 4878367 | 4878368 | 2 | |
| Insertion | Chr6 | 24037839 | 24037842 | 4 | |
| Insertion | Chr9 | 20481962 | 20481963 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 5772842 | 17587064 | |
| Inversion | Chr8 | 13880651 | 13967844 | LOC_Os08g23160 |
| Inversion | Chr8 | 13880881 | 13967864 | LOC_Os08g23160 |
| Inversion | Chr6 | 14150918 | 14477215 | LOC_Os06g24180 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 13880664 | Chr4 | 24094169 | LOC_Os04g40570 |
| Translocation | Chr8 | 13880867 | Chr4 | 24094201 | LOC_Os04g40570 |
| Translocation | Chr12 | 23636397 | Chr5 | 11204746 | 2 |
| Translocation | Chr6 | 29149554 | Chr3 | 27235727 | LOC_Os03g47920 |
| Translocation | Chr6 | 29149555 | Chr3 | 27235728 | LOC_Os03g47920 |
