Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-362 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-362 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 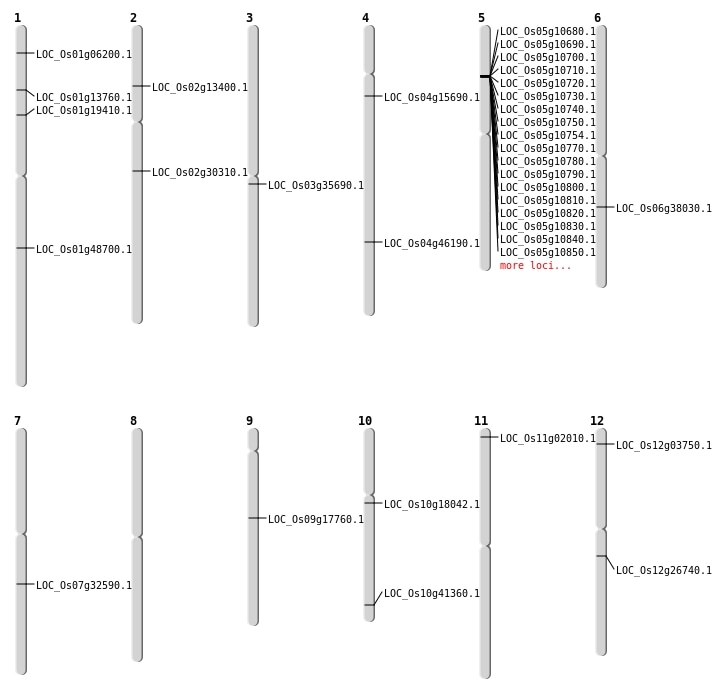
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10982486 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g19410 |
| SBS | Chr1 | 10982487 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g19410 |
| SBS | Chr1 | 30627304 | A-G | HET | INTRON | |
| SBS | Chr1 | 36524177 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 39438429 | G-T | HET | INTRON | |
| SBS | Chr1 | 39438432 | G-T | HET | INTRON | |
| SBS | Chr1 | 39535497 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 39537107 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 40771432 | G-T | HET | INTRON | |
| SBS | Chr10 | 11294952 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 23018680 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 7596014 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 9145015 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g18042 |
| SBS | Chr11 | 24237317 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 28348102 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 12549351 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 18928546 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 23416057 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 18046315 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g30310 |
| SBS | Chr2 | 7145828 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g13400 |
| SBS | Chr3 | 14413091 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 6460815 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 7129959 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 18317969 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 9445444 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 1965199 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 23854540 | G-T | HOMO | INTRON | |
| SBS | Chr5 | 24448941 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 5379369 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 7676719 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 11478896 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 21577017 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 2716436 | G-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 26273248 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 136065 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 18900903 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 5038416 | T-C | HET | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 436512 | 436522 | 10 | |
| Deletion | Chr11 | 537732 | 537742 | 10 | LOC_Os11g02010 |
| Deletion | Chr9 | 901368 | 901370 | 2 | |
| Deletion | Chr12 | 1524981 | 1524992 | 11 | LOC_Os12g03750 |
| Deletion | Chr5 | 5853672 | 7340268 | 1486596 | 201 |
| Deletion | Chr1 | 7369570 | 7369571 | 1 | |
| Deletion | Chr1 | 7712663 | 7712679 | 16 | LOC_Os01g13760 |
| Deletion | Chr5 | 8524430 | 8524435 | 5 | |
| Deletion | Chr4 | 8528720 | 8528722 | 2 | LOC_Os04g15690 |
| Deletion | Chr11 | 8660977 | 8660989 | 12 | |
| Deletion | Chr7 | 8984896 | 8984897 | 1 | |
| Deletion | Chr11 | 9412195 | 9412196 | 1 | |
| Deletion | Chr9 | 10859113 | 10859115 | 2 | LOC_Os09g17760 |
| Deletion | Chr11 | 13186660 | 13186664 | 4 | |
| Deletion | Chr4 | 17203876 | 17203878 | 2 | |
| Deletion | Chr7 | 19026030 | 19026032 | 2 | |
| Deletion | Chr3 | 19769227 | 19769228 | 1 | LOC_Os03g35690 |
| Deletion | Chr4 | 20671766 | 20671769 | 3 | |
| Deletion | Chr1 | 21396308 | 21396326 | 18 | |
| Deletion | Chr5 | 21770001 | 21832000 | 62000 | 9 |
| Deletion | Chr10 | 21963548 | 21963549 | 1 | |
| Deletion | Chr8 | 22082082 | 22082084 | 2 | |
| Deletion | Chr10 | 22512356 | 22512360 | 4 | |
| Deletion | Chr12 | 23535723 | 23535770 | 47 | |
| Deletion | Chr11 | 25755978 | 25755981 | 3 | |
| Deletion | Chr1 | 27927923 | 27928528 | 605 | LOC_Os01g48700 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 42357357 | 42357362 | 6 | |
| Insertion | Chr3 | 32604039 | 32604039 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 5967229 | 6916445 | |
| Inversion | Chr6 | 22434390 | 22481883 | LOC_Os06g38030 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 15670080 | Chr1 | 2952814 | 2 |
| Translocation | Chr7 | 19431518 | Chr5 | 21770265 | LOC_Os07g32590 |
| Translocation | Chr7 | 19432136 | Chr5 | 5853680 | LOC_Os07g32590 |
| Translocation | Chr10 | 22229275 | Chr4 | 27358617 | 2 |
| Translocation | Chr11 | 23286499 | Chr1 | 41587095 |
