Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-411 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-411 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 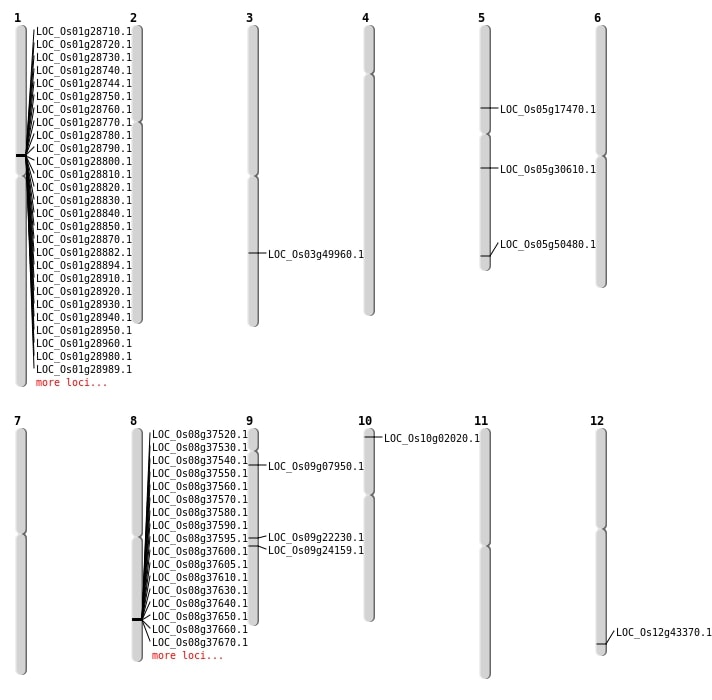
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12445036 | A-G | HET | INTRON | |
| SBS | Chr1 | 25475572 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 31172182 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 32747160 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 39443450 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 644180 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g02020 |
| SBS | Chr11 | 10767908 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 18327659 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 18327660 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 21404841 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 15368557 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 22542881 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 29369078 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 19996278 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 28478898 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g49960 |
| SBS | Chr4 | 13696780 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 26886236 | T-C | HOMO | INTRON | |
| SBS | Chr4 | 27007308 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 35493177 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 5420984 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 8028897 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 10037426 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17470 |
| SBS | Chr5 | 8938715 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 26788379 | A-T | HOMO | INTRON | |
| SBS | Chr6 | 6360577 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 1822603 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 21394611 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 23299744 | A-G | HET | INTRON | |
| SBS | Chr7 | 24782146 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 25233960 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 13447647 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g22230 |
| SBS | Chr9 | 15841345 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 4082641 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07950 |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 256954 | 256967 | 13 | |
| Deletion | Chr5 | 1919262 | 1919278 | 16 | |
| Deletion | Chr10 | 3903197 | 3903199 | 2 | |
| Deletion | Chr7 | 8056484 | 8056485 | 1 | |
| Deletion | Chr2 | 8530750 | 8530773 | 23 | |
| Deletion | Chr2 | 8806079 | 8806080 | 1 | |
| Deletion | Chr9 | 14344742 | 14344743 | 1 | LOC_Os09g24159 |
| Deletion | Chr11 | 14858918 | 14858919 | 1 | |
| Deletion | Chr1 | 16075001 | 16688000 | 613000 | 97 |
| Deletion | Chr5 | 17734265 | 17734277 | 12 | LOC_Os05g30610 |
| Deletion | Chr5 | 20419398 | 20419402 | 4 | |
| Deletion | Chr8 | 23763077 | 23876498 | 113421 | 20 |
| Deletion | Chr4 | 26751968 | 26751969 | 1 | |
| Deletion | Chr5 | 28943960 | 28943961 | 1 | LOC_Os05g50480 |
| Deletion | Chr4 | 29841132 | 29841138 | 6 | |
| Deletion | Chr4 | 30831199 | 30831200 | 1 | |
| Deletion | Chr3 | 31322163 | 31322167 | 4 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 29322201 | 29322201 | 1 | |
| Insertion | Chr10 | 17838567 | 17838570 | 4 | |
| Insertion | Chr12 | 26909980 | 26909982 | 3 | LOC_Os12g43370 |
| Insertion | Chr4 | 20348590 | 20348593 | 4 | |
| Insertion | Chr7 | 4570092 | 4570093 | 2 | |
| Insertion | Chr8 | 22797696 | 22797697 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 16075037 | 42755540 | LOC_Os01g73800 |
| Inversion | Chr1 | 16075492 | 42755533 | LOC_Os01g73800 |
No Translocation
