Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN-95 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN-95 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 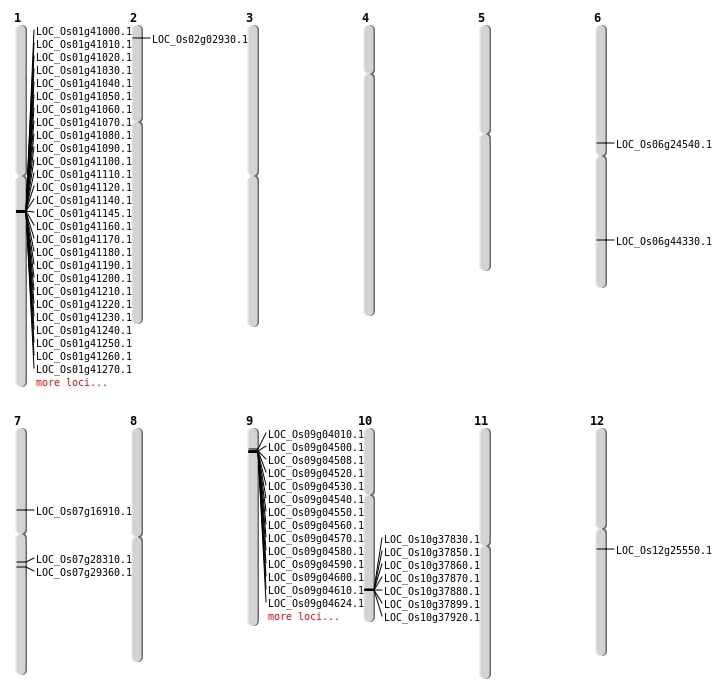
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24698092 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 29336899 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 11105395 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1620832 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 14710659 | A-G | HET | INTRON | |
| SBS | Chr11 | 15206691 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 28840686 | T-A | HOMO | INTRON | |
| SBS | Chr12 | 3708475 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 5370618 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 1146742 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g02930 |
| SBS | Chr2 | 1146743 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g02930 |
| SBS | Chr2 | 1594096 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 17607508 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 24080306 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 29795939 | C-A | HOMO | INTRON | |
| SBS | Chr3 | 29795940 | T-A | HOMO | INTRON | |
| SBS | Chr3 | 34396916 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 11929723 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 21085313 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 28227828 | A-T | HET | INTRON | |
| SBS | Chr4 | 29494664 | T-G | HET | INTRON | |
| SBS | Chr4 | 9457831 | T-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 16438237 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 18959948 | A-G | HET | INTRON | |
| SBS | Chr6 | 13405425 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 14919590 | A-G | HET | INTRON | |
| SBS | Chr7 | 17329279 | T-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 9947749 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16910 |
| SBS | Chr8 | 10019628 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 15350266 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 17771113 | C-T | HET | STOP_GAINED | LOC_Os09g29239 |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1971421 | 1971422 | 1 | |
| Deletion | Chr9 | 2393001 | 2488000 | 95000 | 16 |
| Deletion | Chr5 | 5710045 | 5710048 | 3 | |
| Deletion | Chr3 | 7100740 | 7100752 | 12 | |
| Deletion | Chr12 | 11092824 | 11092825 | 1 | |
| Deletion | Chr1 | 12355812 | 12355854 | 42 | |
| Deletion | Chr6 | 14387413 | 14387438 | 25 | LOC_Os06g24540 |
| Deletion | Chr8 | 15670517 | 15670524 | 7 | |
| Deletion | Chr10 | 15930604 | 15930640 | 36 | |
| Deletion | Chr7 | 16534598 | 16534608 | 10 | LOC_Os07g28310 |
| Deletion | Chr5 | 16807336 | 16807337 | 1 | |
| Deletion | Chr7 | 17237143 | 17237342 | 199 | |
| Deletion | Chr10 | 20248808 | 20311106 | 62298 | 8 |
| Deletion | Chr1 | 23205388 | 24016066 | 810678 | 128 |
| Deletion | Chr2 | 25219647 | 25219664 | 17 | |
| Deletion | Chr4 | 28223159 | 28223168 | 9 | |
| Deletion | Chr3 | 28874021 | 28874039 | 18 | |
| Deletion | Chr3 | 29709718 | 29709726 | 8 | |
| Deletion | Chr1 | 32999641 | 32999643 | 2 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 14786725 | 14786725 | 1 | LOC_Os12g25550 |
| Insertion | Chr3 | 7054743 | 7054743 | 1 | |
| Insertion | Chr3 | 9887960 | 9887960 | 1 | |
| Insertion | Chr7 | 5284335 | 5284337 | 3 | |
| Insertion | Chr8 | 26530008 | 26530008 | 1 | |
| Insertion | Chr9 | 19905483 | 19905488 | 6 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 2059272 | 2392383 | LOC_Os09g04010 |
| Inversion | Chr3 | 27419450 | 28103138 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 2522404 | Chr6 | 26766885 | LOC_Os06g44330 |
| Translocation | Chr12 | 9966059 | Chr7 | 17237343 | LOC_Os07g29360 |
