Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1015-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1015-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 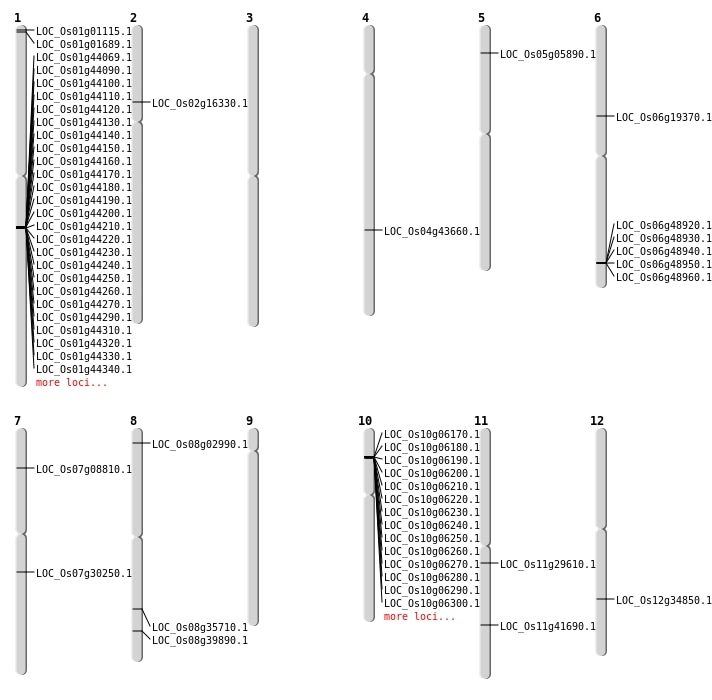
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 1080963 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 17174594 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g29610 |
| SBS | Chr11 | 25032988 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g41690 |
| SBS | Chr11 | 8755327 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 21210817 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g34850 |
| SBS | Chr12 | 23446097 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 23659558 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 426311 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 7238252 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 11039441 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 22765382 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 29689879 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 9288731 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g16330 |
| SBS | Chr3 | 17271090 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 20825427 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 28883911 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 34529598 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr3 | 35276356 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 25833927 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g43660 |
| SBS | Chr5 | 24811987 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 26064374 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 27219533 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 2962415 | G-A | HET | STOP_GAINED | LOC_Os05g05890 |
| SBS | Chr5 | 7147194 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 11024271 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g19370 |
| SBS | Chr6 | 11024272 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g19370 |
| SBS | Chr6 | 14476602 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 5957860 | G-T | HET | INTRON | |
| SBS | Chr7 | 17882978 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30250 |
| SBS | Chr7 | 9426261 | C-T | HET | INTRON | |
| SBS | Chr8 | 1303218 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g02990 |
| SBS | Chr8 | 13163526 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 13163527 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 23816216 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 16342011 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 4680415 | G-T | HET | INTERGENIC |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 3148001 | 3364000 | 216000 | 38 |
| Deletion | Chr5 | 3248952 | 3248953 | 1 | |
| Deletion | Chr7 | 4552171 | 4552177 | 6 | LOC_Os07g08810 |
| Deletion | Chr4 | 4868419 | 4868424 | 5 | |
| Deletion | Chr12 | 17049047 | 17049048 | 1 | |
| Deletion | Chr6 | 20595497 | 20595513 | 16 | |
| Deletion | Chr8 | 22530154 | 22530155 | 1 | LOC_Os08g35710 |
| Deletion | Chr7 | 24712722 | 24712732 | 10 | |
| Deletion | Chr1 | 25241001 | 25604000 | 363000 | 51 |
| Deletion | Chr8 | 25275027 | 25275032 | 5 | LOC_Os08g39890 |
| Deletion | Chr3 | 25970762 | 25970771 | 9 | |
| Deletion | Chr5 | 27548904 | 27548905 | 1 | |
| Deletion | Chr6 | 29614001 | 29670000 | 56000 | 5 |
| Deletion | Chr1 | 33188751 | 33188754 | 3 | LOC_Os01g57420 |
| Deletion | Chr2 | 35214007 | 35214011 | 4 |
Insertions: 8
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 59833 | 352091 | 2 |
| Inversion | Chr1 | 25603645 | 26085728 | LOC_Os01g45120 |
| Inversion | Chr1 | 25824230 | 26085247 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 1340602 | Chr1 | 26085247 | |
| Translocation | Chr8 | 1340605 | Chr1 | 26085250 | |
| Translocation | Chr10 | 3159374 | Chr3 | 8152502 | LOC_Os10g06170 |
| Translocation | Chr12 | 15947415 | Chr4 | 23954007 | |
| Translocation | Chr11 | 15952945 | Chr7 | 11854549 |
