Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1022-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1022-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 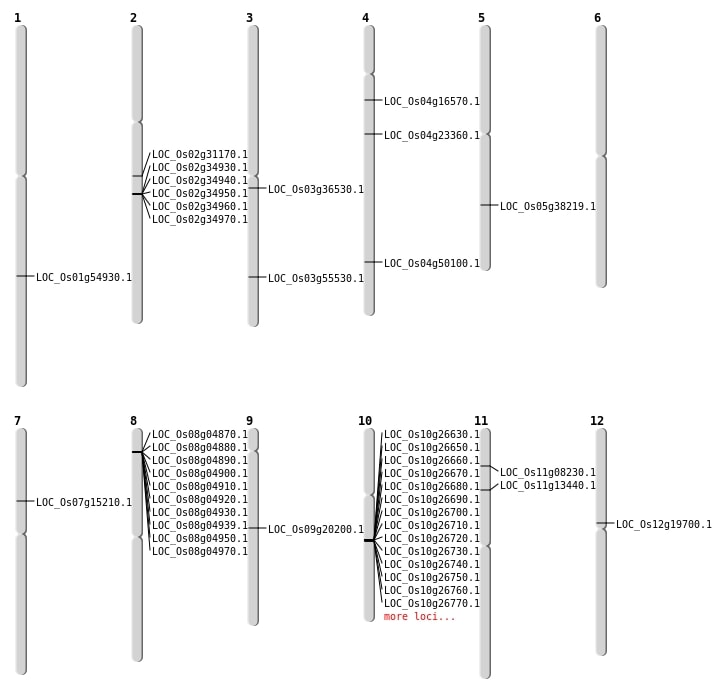
|
| Variant Information |
|---|
Single base substitutions: 61
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14968033 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 18326070 | G-C | HET | INTRON | |
| SBS | Chr1 | 19204175 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 25737710 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 9715775 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 11180781 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr10 | 18925645 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g35400 |
| SBS | Chr11 | 12896705 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 19086600 | G-A | HET | INTRON | |
| SBS | Chr11 | 27741462 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 4323098 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g08230 |
| SBS | Chr11 | 588385 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 7365467 | A-T | HET | STOP_GAINED | LOC_Os11g13440 |
| SBS | Chr12 | 11472904 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g19700 |
| SBS | Chr12 | 12529197 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 12979241 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 11648432 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 30218480 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 33177614 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 34487095 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 3579220 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 7108361 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 13600720 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 19507494 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 2423051 | G-T | HET | INTRON | |
| SBS | Chr3 | 26929899 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 13350602 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g23360 |
| SBS | Chr4 | 16051970 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 20794452 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 21149156 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 22440294 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 23923986 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 33511067 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 34869379 | C-G | HET | UTR_5_PRIME | |
| SBS | Chr4 | 9009536 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g16570 |
| SBS | Chr4 | 9408180 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 14245994 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr5 | 18602499 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 21012802 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 21506239 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 28115451 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 6609905 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 9547274 | T-C | HET | INTRON | |
| SBS | Chr6 | 13641156 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 8021096 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 18276752 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 20140549 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 20374811 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 23382720 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 4217096 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 10370881 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 10774490 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 13937646 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr8 | 1798990 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr8 | 23629541 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 26323932 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 26602117 | T-C | HET | INTRON | |
| SBS | Chr8 | 4115814 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 4356173 | A-T | HET | INTRON | |
| SBS | Chr9 | 20309907 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 3528595 | G-A | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 672949 | 672952 | 3 | |
| Deletion | Chr9 | 1915805 | 1915809 | 4 | |
| Deletion | Chr8 | 2469001 | 2552000 | 83000 | 10 |
| Deletion | Chr8 | 2976757 | 2976763 | 6 | |
| Deletion | Chr7 | 4935320 | 4935321 | 1 | |
| Deletion | Chr1 | 8687820 | 8687824 | 4 | |
| Deletion | Chr7 | 8766011 | 8766023 | 12 | LOC_Os07g15210 |
| Deletion | Chr1 | 10566339 | 10566350 | 11 | |
| Deletion | Chr1 | 10669338 | 10669343 | 5 | |
| Deletion | Chr8 | 10963959 | 10963965 | 6 | |
| Deletion | Chr9 | 12110114 | 12110115 | 1 | LOC_Os09g20200 |
| Deletion | Chr2 | 12483282 | 12483294 | 12 | |
| Deletion | Chr10 | 13910001 | 14293000 | 383000 | 44 |
| Deletion | Chr12 | 14571197 | 14571210 | 13 | |
| Deletion | Chr5 | 17246853 | 17246861 | 8 | |
| Deletion | Chr5 | 18028804 | 18028805 | 1 | |
| Deletion | Chr3 | 20242793 | 20242799 | 6 | LOC_Os03g36530 |
| Deletion | Chr2 | 20952073 | 20973774 | 21701 | 5 |
| Deletion | Chr1 | 28846182 | 28846183 | 1 | |
| Deletion | Chr4 | 29884952 | 29884953 | 1 | LOC_Os04g50100 |
| Deletion | Chr2 | 30176113 | 30176114 | 1 | |
| Deletion | Chr2 | 31187972 | 31187974 | 2 | |
| Deletion | Chr1 | 31596999 | 31597014 | 15 | LOC_Os01g54930 |
| Deletion | Chr3 | 31599808 | 31599810 | 2 | LOC_Os03g55530 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 25174117 | 25174118 | 2 | |
| Insertion | Chr2 | 18667295 | 18667295 | 1 | LOC_Os02g31170 |
| Insertion | Chr3 | 22313537 | 22313539 | 3 | |
| Insertion | Chr4 | 351665 | 351666 | 2 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 2084274 | 2112217 | |
| Inversion | Chr8 | 2084286 | 2468705 | |
| Inversion | Chr8 | 2100661 | 3004339 | |
| Inversion | Chr8 | 2551984 | 3004346 | |
| Inversion | Chr10 | 13898132 | 14763341 | LOC_Os10g26630 |
| Inversion | Chr2 | 20952591 | 21164650 | |
| Inversion | Chr2 | 20973214 | 21163980 | LOC_Os02g34970 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2084270 | Chr1 | 24677418 | |
| Translocation | Chr4 | 12914757 | Chr2 | 20952485 | |
| Translocation | Chr4 | 12914771 | Chr2 | 20952075 | |
| Translocation | Chr10 | 13898135 | Chr2 | 17673926 | LOC_Os10g26630 |
| Translocation | Chr10 | 13910161 | Chr2 | 20973219 | LOC_Os02g34970 |
| Translocation | Chr10 | 14292556 | Chr2 | 21163983 | |
| Translocation | Chr10 | 14763351 | Chr2 | 21164684 | |
| Translocation | Chr5 | 22411100 | Chr1 | 28240038 | LOC_Os05g38219 |
| Translocation | Chr5 | 22411102 | Chr1 | 28241032 | LOC_Os05g38219 |
