Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1024-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1024-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 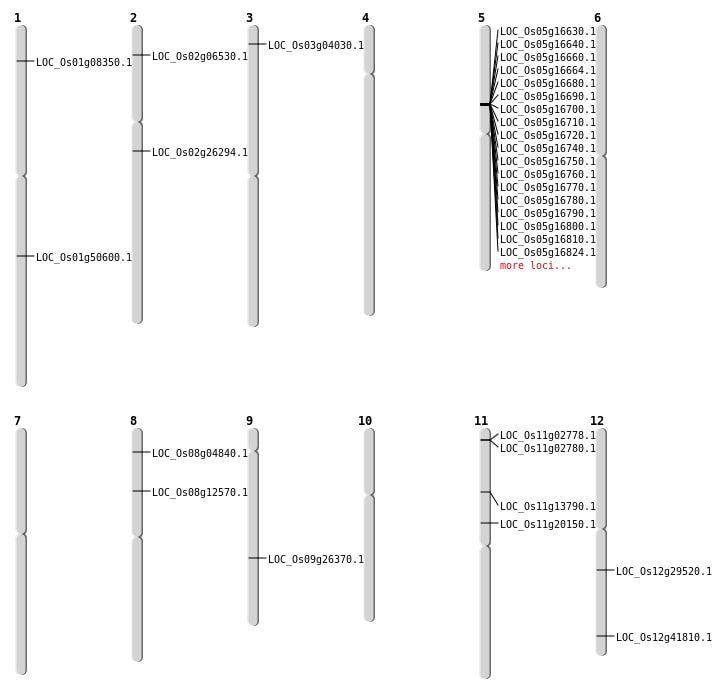
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16300647 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 17538979 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 18144472 | C-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 18369455 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 5384481 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 581000 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 11743618 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 15038327 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 16561166 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 17033949 | A-G | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 3166337 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 17856271 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 20908358 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 8344715 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 25801249 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 16653387 | G-A | HET | INTRON | |
| SBS | Chr2 | 7352560 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 14892586 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 1838424 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g04030 |
| SBS | Chr3 | 20786702 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 23030206 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 3327118 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 36091007 | C-T | HET | INTRON | |
| SBS | Chr4 | 30540439 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 10063255 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 11407881 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 25744401 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 7501555 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 20211010 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 25778143 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 28903701 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 11138938 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 18745419 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 19173644 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 15185447 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 8612390 | T-A | HET | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1203597 | 1203935 | 338 | |
| Deletion | Chr8 | 1203764 | 1203765 | 1 | |
| Deletion | Chr10 | 2165624 | 2165626 | 2 | |
| Deletion | Chr2 | 3264962 | 3264965 | 3 | LOC_Os02g06530 |
| Deletion | Chr7 | 3623670 | 3623678 | 8 | |
| Deletion | Chr5 | 3644599 | 3644679 | 80 | |
| Deletion | Chr1 | 4095879 | 4095887 | 8 | LOC_Os01g08350 |
| Deletion | Chr1 | 6776486 | 6776487 | 1 | LOC_Os01g12400 |
| Deletion | Chr2 | 6795209 | 6795213 | 4 | |
| Deletion | Chr11 | 7580726 | 7580743 | 17 | LOC_Os11g13790 |
| Deletion | Chr11 | 7819774 | 7819780 | 6 | |
| Deletion | Chr12 | 8546932 | 8546933 | 1 | |
| Deletion | Chr5 | 9434001 | 11024000 | 1590000 | 222 |
| Deletion | Chr10 | 9514978 | 9514981 | 3 | |
| Deletion | Chr9 | 13152221 | 13152233 | 12 | |
| Deletion | Chr2 | 15445819 | 15445820 | 1 | LOC_Os02g26294 |
| Deletion | Chr5 | 15804062 | 15804073 | 11 | |
| Deletion | Chr12 | 17574050 | 17574067 | 17 | LOC_Os12g29520 |
| Deletion | Chr1 | 18829359 | 18829361 | 2 | |
| Deletion | Chr7 | 19164358 | 19164365 | 7 | |
| Deletion | Chr1 | 19371433 | 19371505 | 72 | |
| Deletion | Chr2 | 19855033 | 19855034 | 1 | |
| Deletion | Chr3 | 20076228 | 20076283 | 55 | |
| Deletion | Chr5 | 23579147 | 23579148 | 1 | |
| Deletion | Chr8 | 24914152 | 24914177 | 25 | |
| Deletion | Chr11 | 25109812 | 25109816 | 4 | |
| Deletion | Chr12 | 25895809 | 25895813 | 4 | LOC_Os12g41810 |
| Deletion | Chr5 | 27188430 | 27188431 | 1 | LOC_Os05g47450 |
| Deletion | Chr2 | 27346941 | 27346957 | 16 | |
| Deletion | Chr2 | 27484520 | 27484575 | 55 | |
| Deletion | Chr1 | 29049966 | 29049975 | 9 | LOC_Os01g50600 |
| Deletion | Chr2 | 32743086 | 32743103 | 17 | |
| Deletion | Chr3 | 33477732 | 33477733 | 1 | |
| Deletion | Chr2 | 33868457 | 33868458 | 1 |
Insertions: 10
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 916717 | 925830 | 2 |
| Inversion | Chr8 | 2427433 | 2435528 | LOC_Os08g04840 |
| Inversion | Chr12 | 12448502 | 24680510 | |
| Inversion | Chr9 | 15877958 | 15927800 | LOC_Os09g26370 |
| Inversion | Chr2 | 17992504 | 18258774 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1222901 | Chr8 | 16258229 | |
| Translocation | Chr5 | 3089001 | Chr2 | 16394858 | |
| Translocation | Chr2 | 7954256 | Chr1 | 15637541 | |
| Translocation | Chr11 | 11537183 | Chr8 | 7441757 | LOC_Os08g12570 |
| Translocation | Chr11 | 11579450 | Chr2 | 27655695 | |
| Translocation | Chr11 | 11643136 | Chr2 | 27655651 | LOC_Os11g20150 |
| Translocation | Chr11 | 11643228 | Chr3 | 15725439 | LOC_Os11g20150 |
| Translocation | Chr9 | 20287708 | Chr6 | 22062273 |
