Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1026-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1026-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 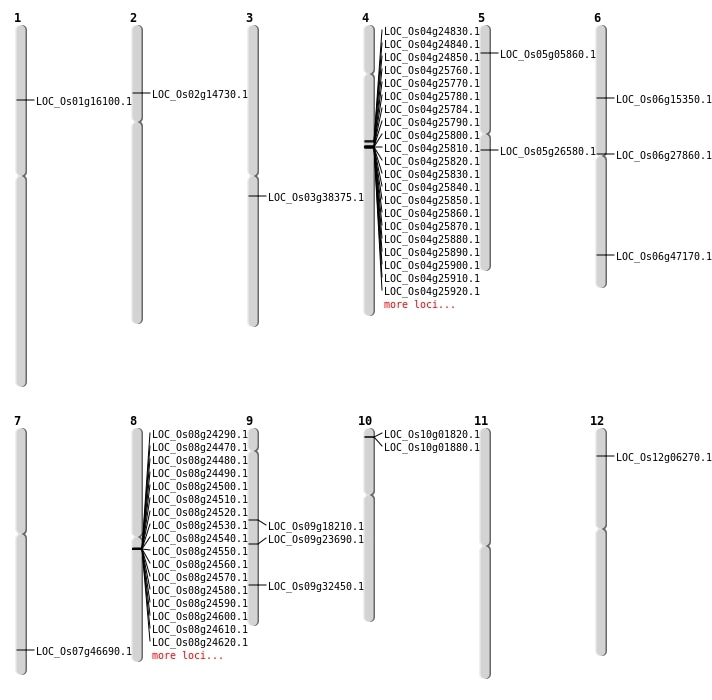
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31248985 | C-A | HET | INTRON | |
| SBS | Chr1 | 37984040 | G-A | HET | INTRON | |
| SBS | Chr10 | 16607758 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 20000424 | C-T | HET | INTRON | |
| SBS | Chr10 | 567693 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g01880 |
| SBS | Chr12 | 17213776 | A-T | HET | INTRON | |
| SBS | Chr2 | 11173006 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 12095564 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 14294313 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 26107462 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 23269234 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 34352986 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 14317018 | A-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 14940182 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 14940183 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 14541297 | G-T | HOMO | INTRON | |
| SBS | Chr5 | 15442838 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g26580 |
| SBS | Chr5 | 15442839 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g26580 |
| SBS | Chr5 | 23389221 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 3232795 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr6 | 25520915 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 28604747 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g47170 |
| SBS | Chr7 | 12546638 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 20164188 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 23272887 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 27911963 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g46690 |
| SBS | Chr7 | 6890374 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 10351905 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 11142083 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 20378470 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 22757082 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 11162605 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g18210 |
| SBS | Chr9 | 11298818 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 13805119 | C-G | HOMO | INTRON |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1615511 | 1615512 | 1 | |
| Deletion | Chr12 | 2993061 | 2993073 | 12 | LOC_Os12g06270 |
| Deletion | Chr6 | 4758682 | 4758719 | 37 | |
| Deletion | Chr1 | 5847464 | 5847465 | 1 | |
| Deletion | Chr5 | 6279216 | 6279219 | 3 | |
| Deletion | Chr2 | 7380757 | 7380758 | 1 | |
| Deletion | Chr6 | 8715184 | 8715185 | 1 | LOC_Os06g15350 |
| Deletion | Chr1 | 9069092 | 9069094 | 2 | LOC_Os01g16100 |
| Deletion | Chr5 | 13475446 | 13475448 | 2 | |
| Deletion | Chr4 | 14268001 | 14314000 | 46000 | 3 |
| Deletion | Chr8 | 14781001 | 14887000 | 106000 | 17 |
| Deletion | Chr12 | 14880557 | 14880558 | 1 | |
| Deletion | Chr4 | 14940001 | 15193000 | 253000 | 35 |
| Deletion | Chr6 | 15785801 | 15785805 | 4 | LOC_Os06g27860 |
| Deletion | Chr11 | 15865780 | 15865800 | 20 | |
| Deletion | Chr8 | 16632332 | 16632358 | 26 | LOC_Os08g27240 |
| Deletion | Chr9 | 19369327 | 19369328 | 1 | LOC_Os09g32450 |
| Deletion | Chr8 | 19563169 | 19563186 | 17 | |
| Deletion | Chr1 | 20092257 | 20092258 | 1 | |
| Deletion | Chr6 | 21111909 | 21111913 | 4 | |
| Deletion | Chr3 | 21303494 | 21303495 | 1 | LOC_Os03g38375 |
| Deletion | Chr8 | 21718206 | 21718210 | 4 | |
| Deletion | Chr10 | 22264559 | 22264866 | 307 | |
| Deletion | Chr1 | 27084006 | 27084016 | 10 | |
| Deletion | Chr5 | 27961822 | 27961824 | 2 | |
| Deletion | Chr5 | 28297593 | 28297594 | 1 |
Insertions: 6
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 533125 | 590023 | LOC_Os10g01820 |
| Inversion | Chr10 | 533138 | 590022 | LOC_Os10g01820 |
| Inversion | Chr8 | 10785698 | 10885645 | |
| Inversion | Chr5 | 14325403 | 14327669 | |
| Inversion | Chr4 | 14940002 | 15225349 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2636510 | Chr2 | 8137782 | LOC_Os02g14730 |
| Translocation | Chr5 | 2937970 | Chr4 | 15231989 | 2 |
| Translocation | Chr6 | 4640248 | Chr1 | 5999921 | |
| Translocation | Chr11 | 5320383 | Chr4 | 14950536 | |
| Translocation | Chr6 | 5737627 | Chr1 | 5999629 | |
| Translocation | Chr9 | 14083782 | Chr8 | 14660870 | LOC_Os08g24290 |
| Translocation | Chr9 | 14087193 | Chr8 | 14886734 | 2 |
| Translocation | Chr10 | 22264582 | Chr1 | 5999629 | |
| Translocation | Chr10 | 22264583 | Chr6 | 4640226 | |
| Translocation | Chr12 | 25546509 | Chr8 | 2637767 | |
| Translocation | Chr12 | 25547630 | Chr2 | 8137784 | LOC_Os02g14730 |
