Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1042-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1042-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 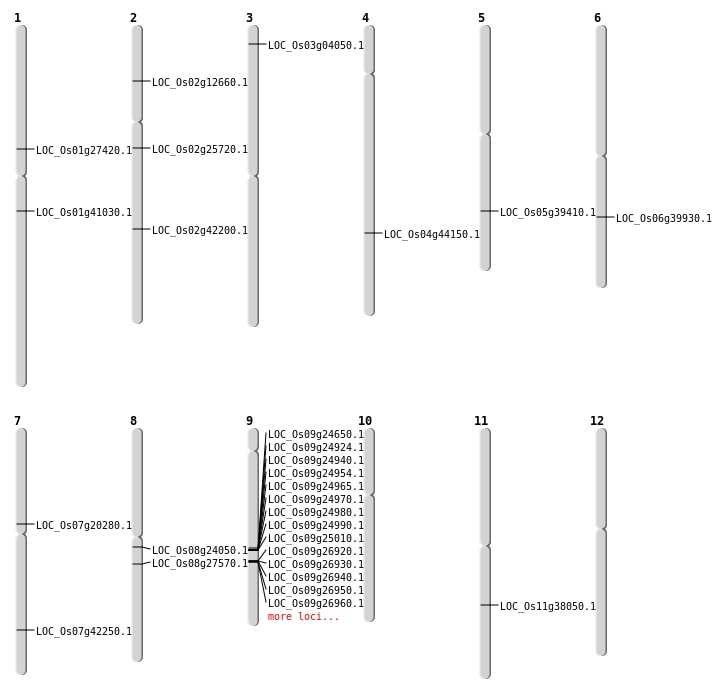
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12065891 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 15304493 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g27420 |
| SBS | Chr1 | 18788652 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 19759768 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 2027345 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 25371395 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 39822307 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr10 | 16062725 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 2334864 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3069754 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 15355429 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 16119841 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 25702674 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 14241212 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 10199560 | T-A | HET | INTRON | |
| SBS | Chr2 | 13544544 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 15062622 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g25720 |
| SBS | Chr2 | 20529326 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 24050699 | T-A | HET | INTRON | |
| SBS | Chr2 | 24050700 | T-A | HET | INTRON | |
| SBS | Chr2 | 30387239 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 30387240 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 35132292 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 6611548 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g12660 |
| SBS | Chr3 | 16765578 | A-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 21564262 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 14865764 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 15038053 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 19486841 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 20150305 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 25276931 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 25601874 | G-T | HET | INTRON | |
| SBS | Chr4 | 4604840 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 20369899 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 23114199 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g39410 |
| SBS | Chr5 | 23208358 | T-G | HOMO | INTRON | |
| SBS | Chr5 | 7728016 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 2348079 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 23749819 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g39930 |
| SBS | Chr6 | 3735538 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 3738806 | T-A | HET | INTRON | |
| SBS | Chr6 | 7179726 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 11692327 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g20280 |
| SBS | Chr7 | 19022988 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 21582576 | A-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 13754419 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 16996960 | G-A | HET | INTRON |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1853005 | 1853007 | 2 | LOC_Os03g04050 |
| Deletion | Chr11 | 9920830 | 9920831 | 1 | |
| Deletion | Chr6 | 14606073 | 14606090 | 17 | |
| Deletion | Chr1 | 14747424 | 14747430 | 6 | |
| Deletion | Chr9 | 14884001 | 14931000 | 47000 | 7 |
| Deletion | Chr9 | 14943001 | 14955000 | 12000 | LOC_Os09g25010 |
| Deletion | Chr9 | 16356001 | 16416000 | 60000 | 8 |
| Deletion | Chr3 | 17068788 | 17068795 | 7 | |
| Deletion | Chr11 | 22940302 | 22940303 | 1 | |
| Deletion | Chr1 | 23234977 | 23234987 | 10 | LOC_Os01g41030 |
| Deletion | Chr4 | 24510717 | 24510718 | 1 | |
| Deletion | Chr8 | 27928794 | 27928803 | 9 |
No Insertion
Inversions: 10
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 14543813 | 16806724 | 2 |
| Inversion | Chr9 | 14675430 | 14883585 | LOC_Os09g24650 |
| Inversion | Chr9 | 14675476 | 14955479 | LOC_Os09g24650 |
| Inversion | Chr9 | 14931387 | 16458441 | 2 |
| Inversion | Chr9 | 14954927 | 18511916 | LOC_Os09g30411 |
| Inversion | Chr9 | 15293442 | 16458438 | LOC_Os09g27050 |
| Inversion | Chr4 | 26134122 | 26629335 | LOC_Os04g44150 |
| Inversion | Chr4 | 26629337 | 28137132 | |
| Inversion | Chr7 | 29219184 | 29325144 | |
| Inversion | Chr7 | 29219199 | 29325148 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 7795950 | Chr4 | 16367948 | |
| Translocation | Chr7 | 9228444 | Chr2 | 25382656 | LOC_Os02g42200 |
| Translocation | Chr8 | 9498456 | Chr7 | 25287861 | LOC_Os07g42250 |
| Translocation | Chr12 | 13397036 | Chr9 | 16355281 | |
| Translocation | Chr9 | 14643611 | Chr7 | 2367381 | |
| Translocation | Chr11 | 22570458 | Chr9 | 14955477 | LOC_Os11g38050 |
| Translocation | Chr11 | 22570463 | Chr9 | 14954927 | 2 |
